An interview from Dr. Amy Johnson, Business and Technical Product Manager at Porvair Sciences and Dr. Lewis Francis, Associate Professor at Swansea University.
Please give an overview of epigenetics and the importance of studying factors that impact human epigenetics.
Epigenetics refers to changes or modifications in genetic sequence that occur ‘around’, ‘on top’ or ‘in addition’ to traditional genetics. Several types of epigenetic modifications that can occur, including DNA methylation, histone modifications (acetylation, methylation), nucleosome positioning and RNA-associated silencing. These well characterised epigenetic mechanisms affect the availability of genes to be activated (switched on or off) which in turn impacts gene expression patterns and important cellular processes.
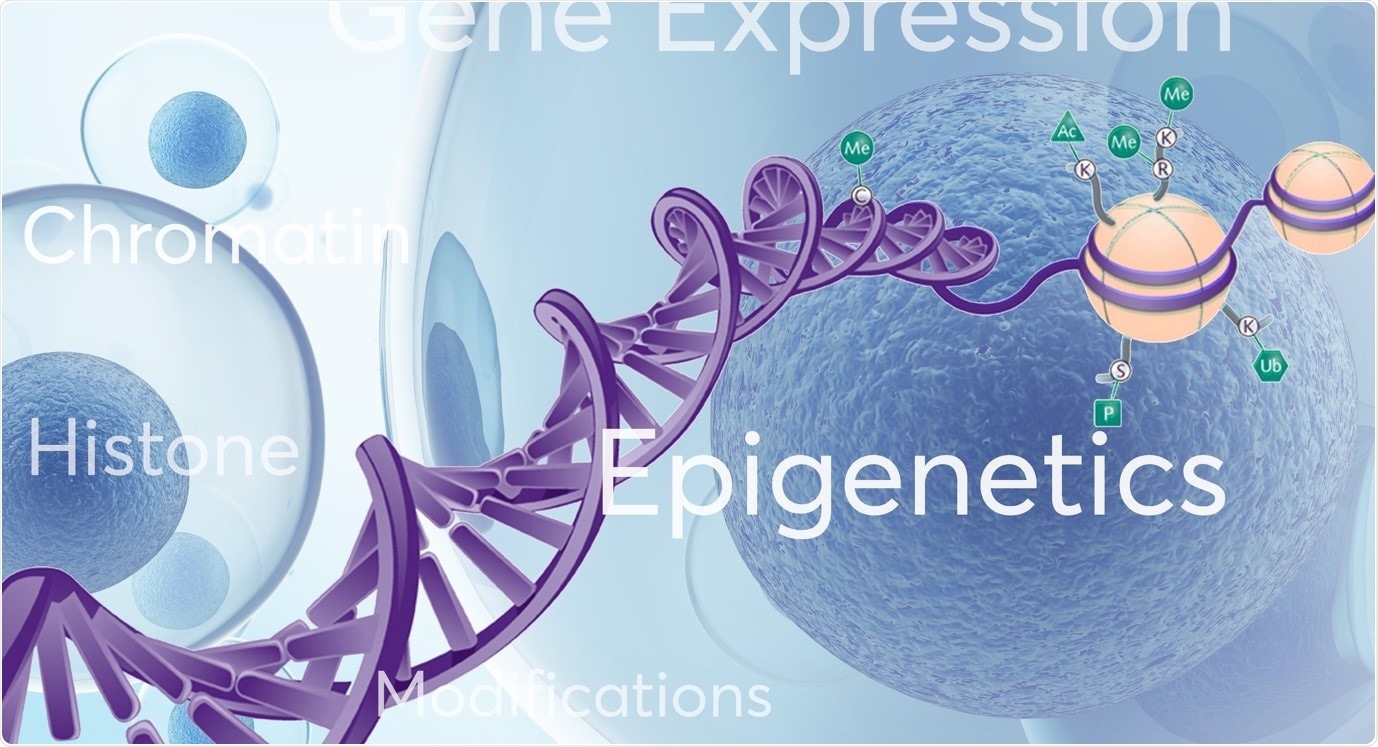
Image credit: Porvair Sciences
Epigenomic regulatory patterns drive cell growth and differentiation mechanisms implicated in development and pathology. Excitingly, technological advances are now enabling genome wide patterns to be deciphered from multi-omics measurements, discovering complex molecular connectivity maps between the genome and its functional output.
Mapping of chromatin accessibility dynamics and higher order chromatin structure has enabled new levels of understanding of cell fate decisions, identity, and function in normal development, physiology, and disease.
How can a virus use epigenetic cofactors to dysregulate cellular pathways and help control its life cycle?
There has been growing evidence that viruses exploit epigenetic processes in order to control their life cycles. The role epigenetics plays in the regulation of viral infections is not yet fully elucidated and the epigenetic regulation of viral gene expression has only recently begun to emerge. Epigenetic regulation during viral infections is usually bidirectional.
Virus will use host cellular factors for transcription or replication and epigenetic cofactors such as histone acetylases, deacetylases, methylases, and demethylases in the control of its life cycle. Viral pathogens have been shown to epigenetically dysregulate cellular pathways in order to optimize their own transcription or replication or evade the cells innate immune response.
For many virus types, nuclear persistence is dependent on genome chromatisation, where virus DNA associates with core histone proteins, forming episomes during latent infection. Integrating viruses such as retroviruses and lentiviruses insert their DNA into host genomes, facilitating histone associations so that the viral chromatin is then subject to histone modifications. This viral epigenome, like the host cell machinery, has significant impact on viral gene expression and replication.
Image credit: Porvair Sciences
A large family of viruses, human papillomaviruses contain a circular, double-stranded DNA genome and its life cycle is dependent on the terminal differentiation of the target cell within epithelia —the keratinocyte.
The virus life cycle begins in basal epithelia, with viral chromatin maintained in an epigenetically repressed state, stabilized by distal chromatin interactions between the viral enhancer and early gene region.
Migration of the infected keratinocyte towards the surface of the epithelium induces cellular differentiation which disrupts chromatin looping and stimulates epigenetic remodelling of the viral chromatin. These epigenetic changes result in enhanced virus transcription and activation of the virus late promoter facilitating transcription of the viral capsid proteins.
What impact can this dysregulation have on cells and organs?
Viruses will disrupt the hosts cell biology and epigenetic processes in order to promote their own replication. This will cause instability in the hosts biological processes by disrupting factors such as DNA replication and transcription and can inhibit pathways of the immune response.
It is important to note that viral infection is not synonymous with disease, as many viral infections are subclinical (i.e., asymptomatic, inapparent), whereas others result in disease of varying severity that is typically accompanied by characteristic clinical signs in the affected host. A virus will infect their host, spread and then damage target tissues.
With a wide variety of strategies to ensure their own survival, there are equally diverse range of associated diseases and pathogenic mechanisms. While viruses differ greatly in their virulence, even a population infected with a single virus will show striking heterogeneity in infection outcomes of individual animals, as shown recently. This can in some way be explained by the wide variety of molecular and cellular impacts.
Infection is often associated with changes in cell morphology, physiology and sequential biosynthetic events – all changes that are necessary for efficient virus replication. From cellular rounding, fusion with surrounding cells, and the formation of cytoplasmic inclusion bodies that represent viral component accumulations, cellular manifestations often result in host cell death.
Cell membrane physiology has been shown to be significantly altered in terms of ionic movements and/or activation cascades leading to altered cellular activities centred on inflammation and metabolism. As outlined here, cellular and nuclear targeting, sometimes through epigenomic processes, often inhibits host cell macromolecular synthesis including DNA, RNA and proteins.
Specific genotoxic effects have also been reported with infection leading to chromosomal breakage, fragmentation or aneuploidy. Dysregulation of such central cell and molecular processes result in significant functional modifications, changing cell shape, growth characteristics, anchorage as well as antigenic or immune properties. In some cases, for both DNA and RNA tumour viruses, they may mediate multiple changes that result in malignant transformation.
Please give an overview of chromatin immunoprecipitation (ChIP).
ChIP is a powerful tool in the study of epigenetics used to uncover associations of specific proteins (modified and unmodified) with defined genomic regions. In a ChIP assay, DNA-protein complexes are selectively immunoprecipitated using matching antibodies and the resulting fractions treated to separate the DNA and protein components. 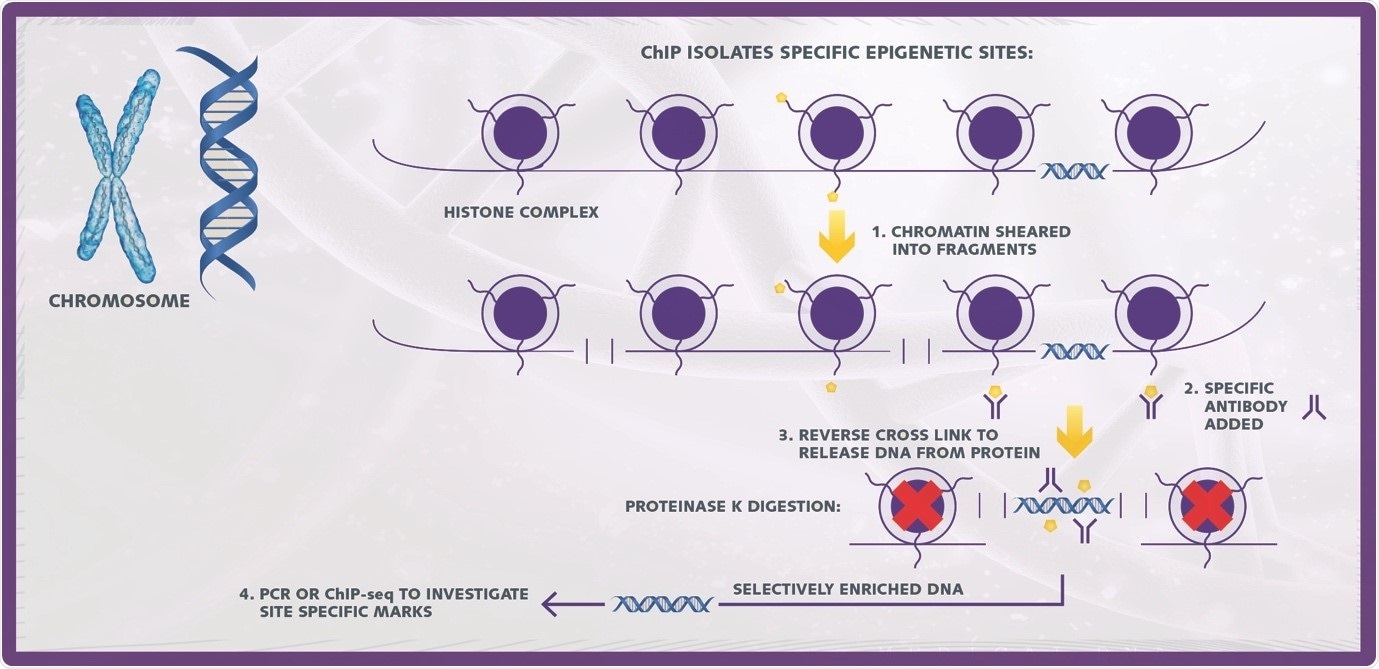
Image credit: Porvair Sciences
A basic ChIP assays consists of 5 key steps
- Chromatin isolation
- Chromatin shearing
- Immunoprecipitation
- Reverse cross linking and DNA purification
- Downstream detection
By using an antibody that is targeting a specific epigenetic regulation in the immunoprecipitation stage and qPCR or next generation sequencing allows investigators to review the epigenetic landscape on a single gene or throughout whole genome.
How can ChIP be used to understand a virus and the impact that a virus has on our epigenetic systems?
The epigenome can differ from cell type to cell type, and within each individual cell, it can potentially modulate gene expression in a number of ways; by organizing the nuclear architecture of the chromosomes, inhibiting or facilitating transcription factor access to DNA, and mediating gene expression.
Deciphering these multifaceted aspects of the epigenome is pivotal for understanding both cell-type-specific gene expression patterns and how viral infection corrupts and utilises host cell epigenetic patterns.
ChIP and ChIP-seq make it possible to determine the location of histones and histone variants linked to viral function alongside the location of proteins of interest such as DNA binding transcription factors and other epigenetic complex proteins to decipher complex regulatory processes that may become hijacked by the virus.
As shown with SV40, better understanding of the mechanisms responsible for the introduction and reading of epigenetic information will lead to novel treatments for viral infections, many of which are serious human pathogens.
Characterizing the mechanisms that underlie each form of epigenetic regulation and dissecting the complex interplay between viral DNA sequences may identify viral proteins and cellular contributors that will lead to new targets for therapeutic intervention.
In short, ChIP assays can locate viral induced epigenetic manipulation related to important cellular functions. Identifying functional epigenetic complexes amenable to therapeutic targeting, the cellular factors that mediate chromatin modification have become attractive target for broad spectrum antiviral therapy, akin to the progress observed in the HIV setting.
What are the advantages and limitations of using ChIP to investigate viruses?
The advantages are that epigenetic marks are well characterised and a wide spectrum of excellent antibodies well characterised. DNA viruses, due to their small sizes, ease of genetic manipulation and the relatively large amounts of chromatin that can be obtained by infections, are efficient targets for ChIP-seq type analysis. With current NGS approaches well established in the pipeline, high, single base resolution, can be achieved allowing subtle alterations in nucleosome location to be identified.
Image credit: Porvair Sciences
As ChIP will only provide information on the epigenetic landscape at a fixed point in time, important consideration however should be given to other omics-based approaches – both DNA/RNA and protein-based to contextualise the chromatin level patterns observed with functional changes in gene expression.
How has ChIP been used to understand the novel coronavirus?
Epigenetic regulation has been the subject of intense interest because of its potential ability to maintain stable gene expression when necessary with the flexibility to respond to changes in environment.
Coronavirus Disease-2019 (COVID-19) respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a novel single-stranded RNA virus of the Coronaviridae family. The disease can be associated with severe respiratory manifestations, cytokine storm, and death.
The emergence of Covid-19 and the variation in progression of the disease amongst the elderly, those with underlying health conditions but also seemingly healthy people is no doubt a combination of several contributing factors where epigenetics may play an important role.
In critical cases of Covid-19, the immune system has been activated and gone into overdrive resulting in the immune system attacking the body’s healthy cells and ultimately leading to cell death and organ failure. The different epigenetic regulation of immune factors such as cytokines could be responsible for certain patients suffering more extreme symptoms of Covid-19.
A recent study published by Sawalha et al., showed overexpression of ACE2 exacerbates the DNA methylation defect in patients suffering with lupus making them more susceptible to severe respiratory complications and death. They determine that inherent epigenetic dysregulation in lupus might facilitate viral entry, viremia, and an excessive immune response to SARS-CoV-2.
How can the results of ChIP help researchers discover and develop therapeutic viral treatments?
There are several suggested clinical management strategies. Once such hypothesis is based on the link between druggable epigenetic modifications and the innate immune response mediated by INFs. IFN pathways are key antiviral response pathways induced early on in infections by cell surface sensing and transcriptional activation of INF stimulated genes as antiviral effectors.
In other viruses such as influenza A, these processes are inactivated using repressive chromatin complexes such as polycomb, which mediates H3k27me3 at antiviral gene promoters. By studying such links in COVID-19, we may identify novel therapeutics strategies, repurposing drugs already in advanced clinical phases for oncological presentations for example.
This is an exciting example of how ChIP mediated identification of epigenomic patterns may play a role in controlling anti-viral response mechanisms in cells. Liberating these newly repressed marks may provide multiple therapeutic approaches. Refer to CEAT project.
Please give an overview of Porvair Sciences’ ChIP product line and the features the ChIP kits that help scientists conduct their research.
Epigenetic analysis is complex, and ChIP often considered a difficult technique due to bottlenecks in speed and throughput. Unlike standard bead-based ChIP assays, Chromatrap® uses a unique filter-based system for the capture and selective enrichment of DNA-associated binding proteins.
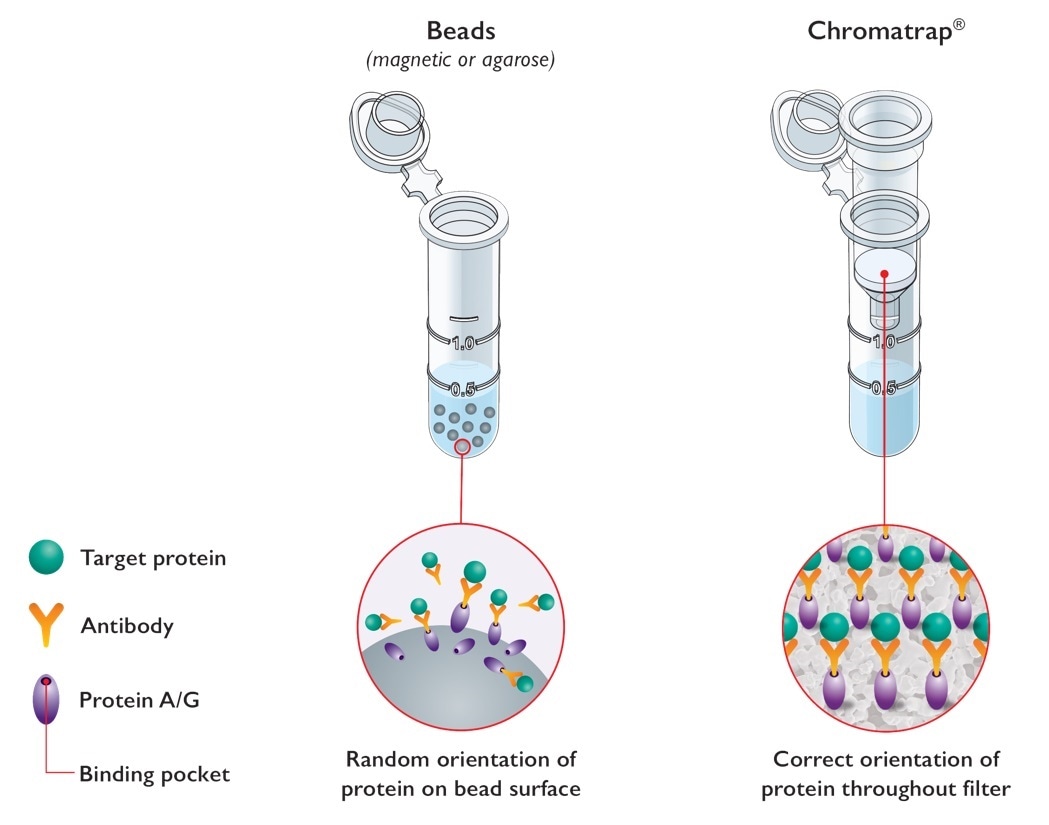
Image credit: Porvair Sciences
By eliminating beads (magnetic and agarose) from our ChIP technology, we were able to develop a ChIP assay that is far easier, quicker, more robust and most importantly more sensitive than standard ChIP assays.
Offered in both a centrifugation spin column format and high throughput 96 well plate we target those bottlenecks providing an easier, faster and more sensitive ChIP. The high throughput format of Chromatrap ChIP makes it an ideal system for personalised medicine applications.
What is the future of ChIP and the study of virus epigenetics?
ChIP can play an important part in understanding the role of epigenetic regulation in maintaining viral episomes through the generation of chromatin, temporally controlling transcription from viral genes during the course of an infection, regulating latency and the switch to a lytic infection, and global dysregulation of cellular function.
By understanding the mechanisms through which viral pathogens manipulate host chromatin, we will understand new aspects of ubiquitous viruses and shed light on previously unknown aspects of chromatin biology. Importantly, by studying the molecular maps of histone marks associated with open and closed chromatin we can target enzyme complexes recruited by the virus. These then become therapeutic targets, in attempts to revert the epigenetic changes to the host genome.
The development of specific inhibitors has tremendous potential for new classes of antivirals and new promise of novel approaches to reducing or eliminating latent viral pools. At present, model systems have clearly demonstrated the potential of these approaches.
However, concerted efforts to recognize and drive these approaches to clinical therapies are needed, with advances in ChIP technology used to track target marks and enzyme complexes and monitor therapeutic efficacy and mechanism.
As there have been significant advances in the development of inhibitors or other chromatin modulation compounds for oncology, the parallels of chromatin modulation in viral diseases should be clearly recognized and considered.
Inflammation and metabolism are exciting avenues based upon chromatin-mediated regulation of the cellular antiviral response and control of inflammation. Modulation of viral infection by chromatin involves the coordinated scientific assessment of the epigenomes of various viral families.
It is likely that ChIP assays will continue to provide genome wide datasets and databases, capturing the state of viral chromatin, enabling additional regulatory circuits to be better understood with respect to variances between viral states, cell-type dependence and specific modulation components. Intense efforts will then allow a range of compounds to be identified, fabricated or re-purposed toward targeting these chromatin regulatory and recognition components in virology.
Where can readers find more information?
https://www.microplates.com/chromatrap/

About Dr. Amy Johnson
Dr. Amy Johnson leads the business and technical development at Porvair Sciences. Shortly after gaining her PhD in Neuroscience from Swansea University, Dr. Johnson established the development of the Chromatrap technology and product range for the epigenetics market. Together with a leading group of scientists and engineers, Dr. Johnson continues to focus on developing new technologies and solutions for sample preparation and life science applications.
About Dr. Lewis Francis

Dr Francis is an Associate Professor in Swansea University, exploring cellular heterogeneity in complex tissue micro-environments. Lewis’ research aims to identify distinct cellular and molecular networks for therapeutic targeting, focusing on the exploration of biomarker expression, gene regulation networks and cell-matrix interactions in niche tissue spaces.