Apr 25 2018
A new tool from the HBP helps to investigate the role of genes in disease-relevant brain areas
Elucidating the way from genes to cognition requires detailed insight into how anatomical areas with specific gene activities contribute to brain function and dysfunction. Up to now, the scientific community has developed many separate brain atlases for different modalities and scales. Now, a team of researchers from the European Human Brain Project (HBP) has developed the JuBrain Gene Expression tool (JuGEx) that combines the benefits of a genetic and an anatomical atlas. Using JuGEx, the scientists found differentially expressed candidate genes for major depressive disorder in a disease-affected part of the brain.
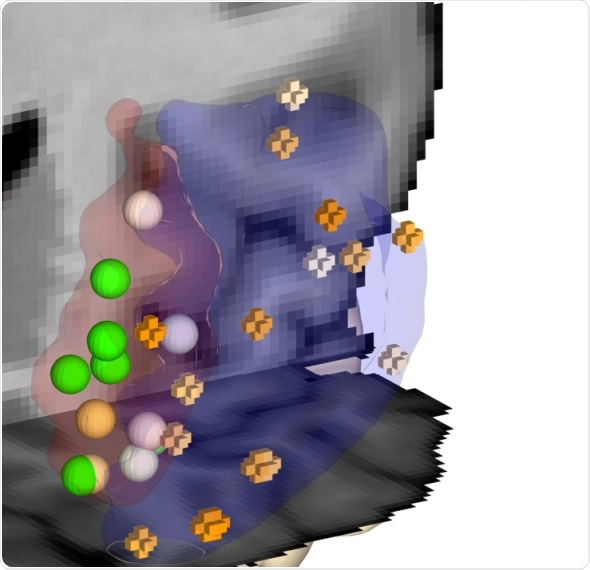
The medial frontal pole (red) is affected in major depression. JuGEx allows to locate gene expression data from tissue samples (colored spheres) ©Forschungszentrum Jülich / HBP
To describe the complexity of human brains, a number of large-scale mapping efforts have been initiated around the world resulting in individual atlases which focus on particular aspects. At the level of genetics and anatomy, two atlases are well-acknowledged: The American Allen Brain Atlas is a resource for gene expression and the German JuBrain Atlas provides maps of cell architecture, reflecting the inter-individual variability between brains.
“Linking the data that have been produced by both atlases would be a milestone to bridge between scales and modalities in our understanding of brain organization. We need to find ways of tracking how changes at one level translate to the next level”, says Katrin Amunts. The neuroscientist is the Scientific Director of the HBP and leads the development of the HBP Atlas, a digital 3D model that integrates many different scales and modalities.
The team has now launched JuGEx which allows to relate genetic data from Allen Brain to the microstructural maps of JuBrain. “Our approach makes it easy to test for differential gene expression between freely selectable pairs of brain areas”, says Thomas Mühleisen, a geneticist at Forschungszentrum Jülich in Germany. Together with Sebastian Bludau, a neuroanatomist at the same institute, he led the development of JuGEx. “If you are interested in a brain feature that has been identified by structural magnetic resonance imaging, for instance, gene expression in this region can now be explored with high spatial precision”, Mühleisen explains. JuGEx will be extended in the next weeks and months, together with Michael Hawrylycz, collaborator from the Allen Brain Institute in Seattle.
To demonstrate the added value of JuGEx, the team chose the topic of major depressive disorder (MDD), a heritable psychiatric disease. "In one of our imaging studies with JuBrain maps, we had found that grey matter volume in the so-called medial frontal pole is significantly reduced in patients with this pathology compared to healthy controls" explains Bludau. “This area is microstructurally distinct and involved in emotion regulation.” Then the team used JuGEx to investigate a bunch of candidate genes for MDD in this area and got two hits: A significant upregulation of the genes MAOA and TAC1. MAOA encodes an enzyme involved in the degradation of neurotransmitters like serotonin, and TAC1 generates hormones that play a role in neuron excitations and behavioral responses.
"As with other tools of the HBP Atlas, JuGEx will be continuously developed further and will receive added functionality. We hope that a broader community will find it useful for their work", Katrin Amunts concludes.