Feb 13 2019
The new Deeplex®-MycTB mycobacterial antibiotic resistance test, developed by GenoScreen, has been used by an international research team in a molecular epidemiology study published in The Lancet Infectious Diseases. This study highlights the spread of tuberculosis strains resistant to first-line antibiotics, not detected by actual international tests.
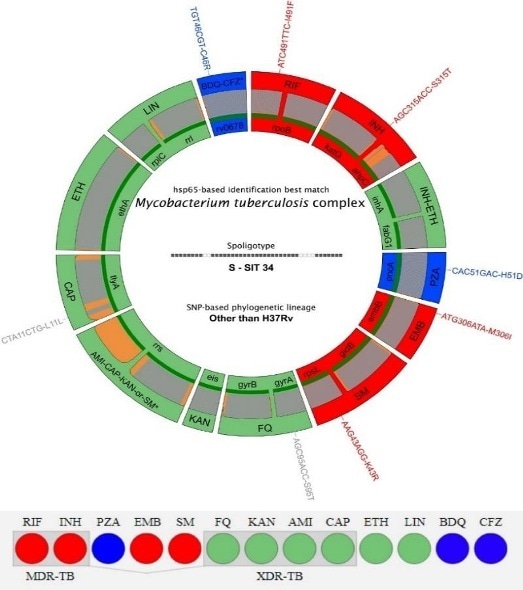
1: Visualisation and interpretation of Deeplex®-MycTB results.
The circle, or "Deeplex map", represents the genes studied with their names on the inside and the antibiotic(s) concerned on the outside.
Color refers to the results of antibiotic susceptibility prediction (red for resistant, green for sensitive and blue for "not characterised until now in the scientific literature").
Mutations are indicated on the outside of the circle, using the same colour code (grey for mutations without effect on resistance).
The resistance results are synthesised by a "resistotype" shown below, using the same color codes associated with the resistance prediction for each antibiotic (RIF, INH, PZA...).
With 10 million new cases per year and 1.6 million deceases in 2017, tuberculosis is the most deadly infectious disease in the world. The number of new cases of multidrug-resistant tuberculosis emerging each year (estimated at 450,000 new cases in 2017) is a global public health problem.
On October 18th 2018, a study conducted in southern Africa, co-directed by Dr P. SUPPLY (CNRS/CHU/INSERM/ The University of Lille /Institut Pasteur de Lille), Dr B. de JONG (Institute of Tropical Medecine of Antwerp) and Dr E. ANDRÉ (Université Catholique de Louvain), has been published in The Lancet Infectious Diseases. It reveals that multidrug-resistant strains of tuberculosis are not detected by current diagnostic tests which cause ineffective treatment for patients, increased mortality and contagion, as well as an accumulation of additional resistance in the bacterial strains involved.
These results were achieved, mainly, credits to the innovative molecular test for predicting antibiotic resistance to this disease: the Deeplex®-MycTB test, developed by GenoScreen.
This test is based on the latest DNA sequencing technologies (NGS) and the bioinformatic detection of mutations responsible for antibiotic resistance. A secure web application allows users to access analyze and interpret results using an interactive graphical interface wherever they are. Unlike the culture-specific testing which takes several weeks, the complete sample analyzis can be finalised in just one to three days. After certification (CE-IVD marking) for medical use, Deeplex®-MycTB will be able to help clinicians to define more precisely and more quickly the appropriate treatment for patients.
On the basis of a single molecular test, this test allows to:
- identify more than 140 species of mycobacteria, including Mycobacterium tuberculosis, the agent responsible for tuberculosis,
- predict resistance to more than a dozen antibiotic molecules used in the antituberculosis treatment of this infectious-agent,
- identify the genetic type of the strain involved for epidemiological tracing purposes,
- visualise the results on an interactive graphical interface giving access to the detail of the results.
Contrairement à d’autres tests de détection des résistances, Deeplex®-MycTB présente le triple-avantage d’être rapide, de détecter une large variété de mutations avec une grande sensibilité et de réaliser un typage facile des mycobactéries, pour suivre l’émergence de nouvelles souches résistantes.
[trad : Unlike other resistance detection tests, Deeplex®-MycTB offers the triple advantage of being fast, detecting a wide variety of mutations with high sensitivity and easily typing mycobacteria, to monitor the emergence of new resistant strains.]
Dr. Gaëlle BISCH – « isolated microorganism » R&D Team Manager, GenoScreen."