May 28 2019
Leptospirosis is an emerging zoonotic disease that affects more than one million people around the world each year. Researchers reporting in PLOS Neglected Tropical Diseases have now sequenced the genomes of Leptospira collected from environments around the globe and revealed 30 new species and new patterns of species diversity.
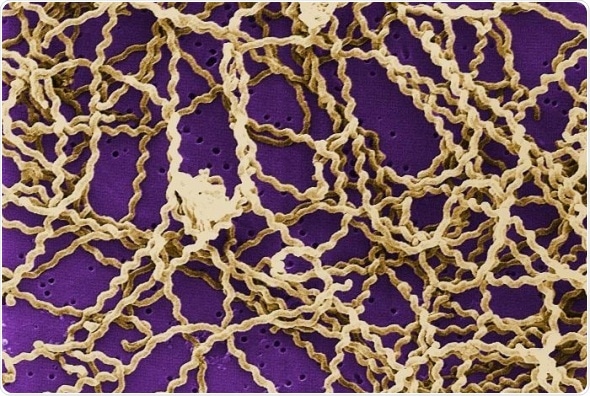
"Leptospira are a member of the phylum Spirochaetes and consist of saprophytic and pathogenic bacteria. Pathogenic species are responsible for the zoonotic disease leptospirosis, where humans end up being an occasional host in a cycle involving wild and domestic animals." Credit: Mathieu Picardeau (CC BY 4.0, 2007)
The genus Leptospira is currently divided into 35 species classified into three phylogenetic clusters that historically correlate with the level of pathogenicity of the species: saprophytic, intermediate, and pathogenic. The evolution of each of these clusters has been unclear and the virulence status of many species is unknown.
In the new work, a group of researchers from the Institut Pasteur International Network (IPIN) including Mathieu Picardeau and Pascale Bourhy, of the Institut Pasteur, France, Frédéric Veyrier of the INRS-Institut Armand-Frappier, Canada, and other colleagues studied 90 Leptospira strains isolated from 18 sites across four continents, including Japan, Malaysia, New Caledonia, Algeria, France, and Mayotte. The genome of each isolate was sequenced and compared to known Leptospira genomes.
Based on the genetic sequences, the team was able to identify 30 new Leptospira species. They organized the Leptospira genus, which now comprises 64 species, into four lineages or subclades, dubbed P1, P2, S1, and S2. The S2 subclade has never been described. In the P1— formerly known as pathogenic—lineage, they shed light on a phenomenon of genome reworking that may explain their evolved pathogenicity.
Researchers say:
We have dusted off the Leptospira genus and gained more clarity of its diversity which will help researchers propose new standards on its classification and nomenclature. The implication of several new potentially infectious Leptospira species for human and animal health remains to be determined but our data also provide new insights into the emergence of virulence in the pathogenic species.”
Source:
PLOS
Journal reference:
Vincent, A.T. et al. (2019) Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLOS Neglected Tropical Diseases. doi.org/10.1371/journal.pntd.0007270.