In this interview, Cedric Uytingco discusses the key benefits of being able to visualize gene expression patterns in neurological samples.
Why is it important for neuroscientists to combine histopathology and gene expression?
Traditionally, neuroscientists used histology to infer cell type based on known morphology and location of cells. However, using this classification alone can obscure information that is necessary to understand normal development and disease pathology.
Gaining an understanding of gene expression patterns and cell-to-cell interactions within tissue provides additional context, magnifying insights that can be gained and accelerating biological discovery.
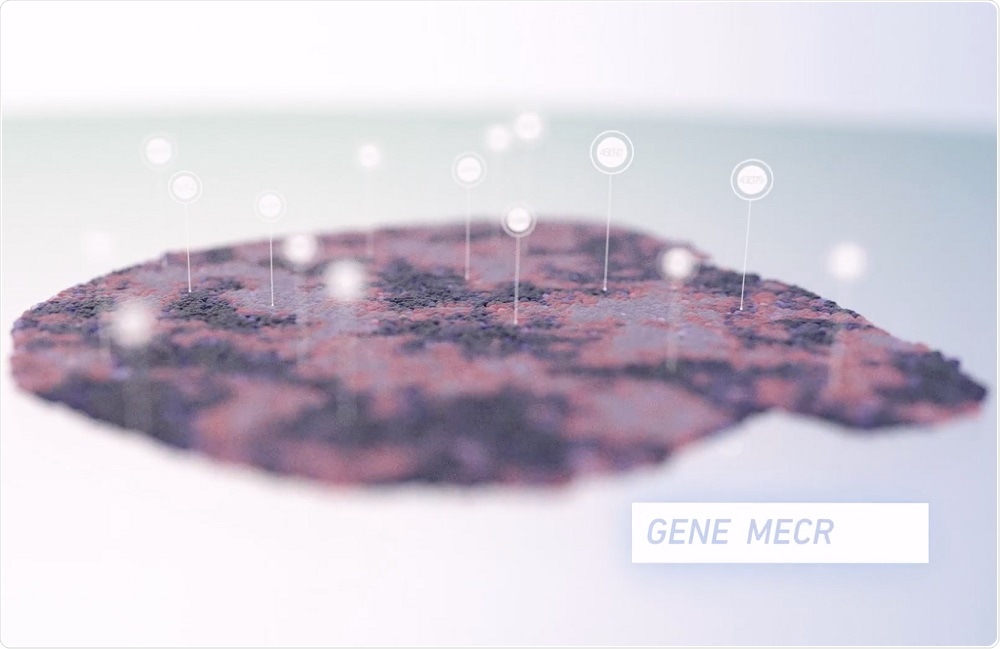 Credit: 10x Genomics
Credit: 10x Genomics
For which other areas of research is this particularly useful?
Connecting gene expression to histology is useful for a wide variety of research applications where researchers want to dive deeper into tissue architecture, biological function, and disease pathology. In addition to neuroscience, researchers studying cancer, immuno-oncology, immunology, and developmental biology will find the technology useful.
What are the main challenges that scientists face when trying to visualize gene expression spatially using traditional methods?
Traditional techniques that examine gene expression and morphology, such as immunohistochemistry or in situ hybridization, rely on probing protein and/or nucleic acids of a relatively small number of known targets. This requires prior knowledge of cell markers and by limiting studies to known targets, researchers may be missing information about cell subtypes that could be biologically significant.
Examining a wide range of biomarkers requires a large number of sections, probed with targets individually or with low-plex throughput, and finally, all the sections need to be imaged and manually analyzed. In cases where the amount of tissue available is limited, analyzing extensive targets may not even be possible.
What is spatial gene expression profiling and how does it overcome some of these issues?
Spatial gene expression profiling provides a view of gene expression patterns by combining histology and total mRNA expression in intact tissue sections. Characterizing cell populations without prior knowledge of cell subtypes or cell markers enables deep insights into gene expression heterogeneity, as well as the discovery of novel biomarkers.
This technology enables a wealth of quantitative information from an individual section, including gene expression patterns, coexpression information, and molecular insights into signaling pathways.
How was the technology developed?
The ability to combine unbiased gene expression data with the histological information from the same tissue section was first described by Ståhl et al. in Science who then founded Spatial Transcriptomics as a spin-off from Science for Life Laboratory, which we acquired in November 2018.
Once part of 10x Genomics, the development effort was intensified to produce a more robust, easy-to-use solution with better spatial resolution, higher sensitivity, and automated analysis tools. Less than a year later 10x Genomics is bringing Visium Spatial Gene Expression Solution to the market.
How does the Visium Spatial Gene Expression Solution work?
Fresh-frozen tissue is sectioned and placed onto a slide with thousands of barcoded spots, containing millions of capture oligonucleotides with spatial barcodes unique to that spot. The tissue sections are imaged and then processed to release mRNA, which binds to the capture oligonucleotides.
A reverse transcription reaction takes place on the slide, generating cDNA that is used to make a standard short-read library. The barcoded cDNA library can be mapped back to a specific spot, creating a layer of gene expression data that can be combined with a microscope image of the tissue section.
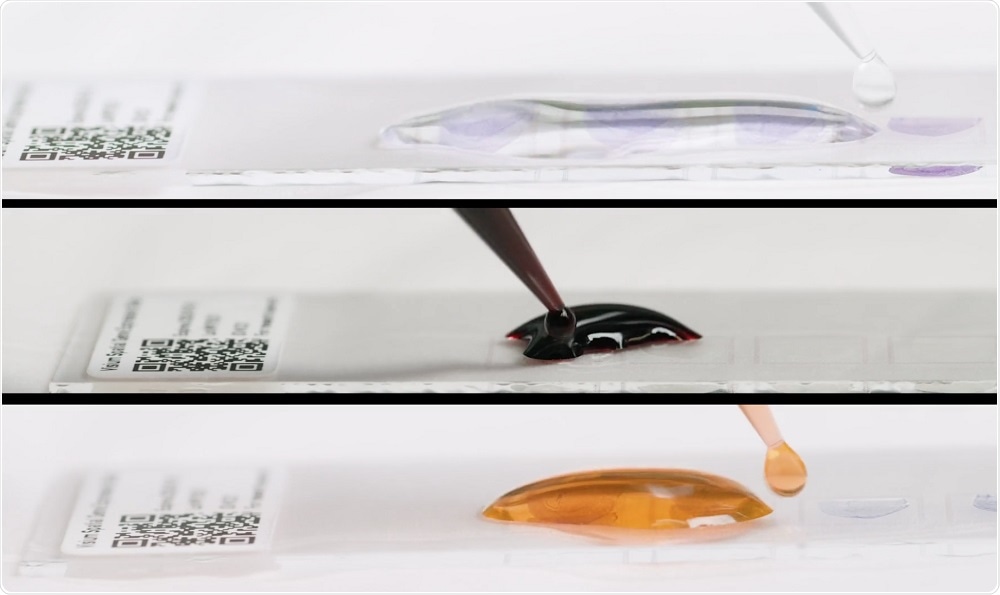 Credit: 10x Genomics
Credit: 10x Genomics
How is spatial gene expression data advancing our understanding of both neurological disorders and the healthy CNS?
The nervous system is immensely complex, developing from a relatively homogenous population of progenitor cells to a diverse network of cells that controls motor and sensory function, as well as cognition. Unraveling the complexities of this system requires not just understanding what is happening but also where, as distinct regions of the nervous system control different functions.
Exploring gene expression patterns in an unbiased manner with spatial gene expression profiling deepens our understanding of the factors that contribute to normal development and function, as well as mechanisms that underlie neural disorders.
The technology was recently used to carry out spatiotemporal gene expression in ALS. Why was this research significant?
The paper by Silas Maniatis and colleagues examined gene expression in postmortem tissue of ALS patients and a murine model of the disease. Their study confirmed alterations in gene expression of several genes known to be involved in ALS but also identified additional changes in genes involved in microglial activation and autophagy.
Additionally, these were able to use morphology to map gene expression changes along the rostrocaudal axis of the spinal cord and between white and gray matter. This research provides insights into the underlying cellular and molecular mechanisms of ALS.
For research in this area and other neurodegenerative diseases, how can this technology play a role?
One major advantage of this technology is the wealth of information that can be gained from a single tissue section, which is particularly advantageous for studies where the amount of tissue is limited.
Researchers will now be able to examine gene expression patterns in tissue and to use the information gained to identify and characterize new biomarkers and signaling pathways that play a role in disease, ultimately leading to a better understanding of disease causes and progression. They can leverage the information gained from these studies to begin to ask questions that previously weren’t possible.
 Credit: CoffeeMill | Shutterstock
Credit: CoffeeMill | Shutterstock
In addition to ALS, how is this technology being used to improve our knowledge of Alzheimer’s disease?
Single-cell RNA sequencing (scRNA-seq) is often used to study the different cell types in the CNS of mammals, but this technique is limited by the dissociation of neurons and glial cells from native tissue. This limits our ability to observe the spatial distribution of gene expression.
This is of particular importance in Alzheimer's research, where we are still trying to identify the contributory roles of various structures in the CNS. At the Society for Neuroscience annual conference, I will be presenting a poster on our studies of spatial gene expression profiling, which will touch on profiling a mouse model of familial Alzheimer’s Disease, APPSWE [Tg2576].
For those who haven’t used this technology before, how difficult is it to incorporate into your research? Does it require specialist equipment or training?
The Visium Spatial Gene Expression Solution is easily adoptable within existing lab infrastructure. It uses standard laboratory methods and tools for tissue analysis, including tissue sectioning, Hematoxylin, and Eosin (H&E) staining, and imaging. The solution is kitted, which includes the slide and reagents necessary to create a sequence-ready library.
Where can our readers find more information?
About Cedric Uytingco, PhD
 Throughout my career, I’ve been focused on neuroscience research and leveraging the information that can be gained from tissue sections to address fundamental biology and its impact on disease.
Throughout my career, I’ve been focused on neuroscience research and leveraging the information that can be gained from tissue sections to address fundamental biology and its impact on disease.
I completed my graduate studies at the University of Maryland, School of Medicine, where my studies focused on functional circuit mapping of the olfactory system.
From there I completed a postdoc at the University of Florida focused on systems biology and examining the pathophysiology of ciliopathies, with the ultimate goal of developing gene therapeutic treatment.
I came to 10x Genomics in January 2019 and joined the team that was focused on developing the Visium Spatial Gene Expression Solution.
About 10x Genomics
10x Genomics is a life science technology company building products to interrogate, understand and master biology to advance human health. The company’s integrated solutions include instruments, consumables, and software for analyzing biological systems at a resolution and scale that matches the complexity of biology.
10x Genomics products have been adopted by researchers around the world including 93 of the top global research institutions and 13 of the top 15 global pharmaceutical companies, and have been cited in over 500 research papers on discoveries ranging from oncology to immunology and auto-immune diseases.
