With modern advances in genetic engineering occurring almost every day, the latest discovery concerns antibiotic resistance. Using the powerful gene editor CRISPR, scientists reported the development of a gene-drive system that is 100 times as efficient as other current systems at inactivating a specific bacterial gene responsible for making the bacterium antibiotic-resistant and which is present as multiple copies within the same bacterial cell. The paper, published on December 16, 2019, in the journal Nature Communications, uses the technology called active genetics, pioneered by biologists at UC San Diego.
Antibiotics have been massively overused over the last few decades of healthcare, all over the world. This period has also seen their routine introduction into animal feed as a part of commercial animal food production. The result of these two trends has been the widespread presence of antibiotics in the environment, both on land and in water. This has in turn caused a high and rising prevalence of antibiotic resistance.
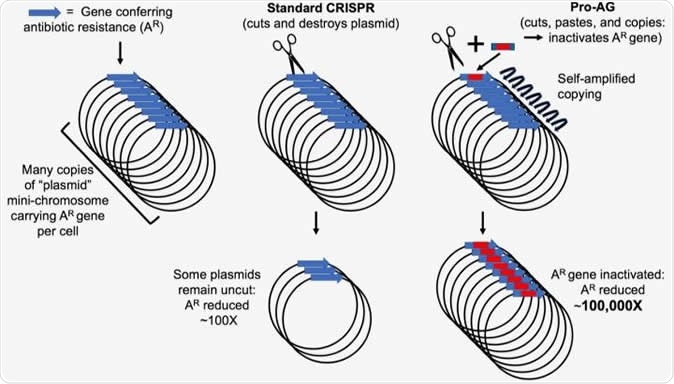
Genes conferring antibiotic resistance (AR) in bacteria (blue arrow) are often carried on circular mini-chromosome elements referred to as plasmids. Site-specific cutting of these plasmids using the CRISPR system, which results in destruction of the plasmid, has been used to reduce the incidence of AR by approximately 100 fold. Pro-Active Genetics (Pro-AG) employs a highly efficient cut-and-paste mechanism that inserts a gene cassette (red box) into the gene conferring AR thereby disrupting its function. The Pro-AG donor cassette is flanked with sequences corresponding to its AR target (blue boxes) to initiate the process. Once inserted into an AR target gene, the Pro-AG element copies itself through a self-amplifying mechanism leading to an approximately 100,000-fold reduction in AR bacteria. Image Credit: Bier Lab, UC San Diego
What is antibiotic resistance?
Antibiotic resistance is the ability of a microbe, whether bacteria, virus or parasite) to prevent the activity of an antibiotic against it, thus allowing the infection to persist and spread to others. The range of bacteria and fungi that are showing antibiotic resistance is increasing by the day. This is a seriously worrying threat to public health and the World Health Organization, as well as many national health organizations, have spelt out measures to contain it and to act against it.
Bacteria acquire antibiotic resistance from other microbes in the environment, and this in turn can be transmitted to humans. As a result, say health professionals, antibiotic resistance is increasing sharply the world over, and in two or three decades, it could go out of hand. They predict that by 2050 there might be about 10 million deaths due to resistant infections, if the current trend is not stopped.
The Pro-AG system
The new technology uses the Pro-AG system, which denotes the pro-active genetic system. This is a method designed to change the way a genetic trait is inherited, in insects and in mammals. These traits are called “preferred traits”. It uses a guide RNA (gRNA) with sequences next to it that are homologous to the targeted site for editing. Once the DNA double strand to be edited is cut, using the gRNA/Cas9 editor, the gRNA is inserted into the break along with any other sequences it carries (the “gRNA cassette”). This makes it a preferred trait so that it is then transmitted to almost every offspring.
To achieve this, the scientists used an adapted form of the CRISPR-Cas9 gene editor to modify DNA from E. coli bacteria. They worked out a method by which they could interrupt the activity of an antibiotic resistance conferring gene. Their main focus is on preventing the transmission of resistance-trait-carrying plasmids.
Plasmids are circular DNA molecules capable of independent replication and not just when the bacterial DNA replicates. This capability means that each plasmid can exist in multiple copies within the bacterial cell. This phenomenon, called plasmid amplification, allows the trait to be transferred from one bacterial strain or species to another, making it very hard to treat more and more infections.
To counter this, the Pro-AG system uses a two-step process to cut out the unwanted material and repair the cut ends. Using this, the scientists were able to achieve results in terms of disrupted antibiotic resistance marker at the target site, at efficiencies that were 100-fold or more that achieved with any comparable method in use – which are, in contrast, based on cutting out and destroying the gene. The edited plasmid is still able to replicate and to spread to other bacteria, but cannot produce drug resistance any more.
To show off the effectiveness of the technique, the researchers used it on bacterial cultures in the laboratory, which had a high number of plasmids that carried known genes conferring resistance to the common antibiotic ampicillin. To do this, the Pro-AG system includes an editing mechanism that amplifies its results, through a positive feedback loop that is activated as the gRNA dose increases with each round of replication. This is a prime reason for its high efficiency.
Once the edited plasmid is ready, it is inserted with extreme precision into the target site. This could one day help patients with chronic bacterial infections to be treated and cured.
The Pro-AG system is as yet in the preclinical stage, but it is not difficult to envisage a refinement that could allow the use of a human delivery system carrying Pro-AG, to eliminate the genetic basis of antibiotic resistance in conditions like cystic fibrosis, tuberculosis, biofilms causing resistant infections, and chronic infections of the urinary tract, which are very challenging in a hospital.
The next step would be to work on combining the Pro-AG system with a range of delivery tools so that it can spread rapidly and widely through multiple populations of resistant bacteria. This could be a very efficient and effective as well as focused way to scrub or remove such antibiotic-resistant traits strains from the bacterium, whether it is from a sewer, a fishpond or a feedlot, according to researcher Ethan Bier.
The unique cut-and-repair mechanism of Pro-AG means that it can also be used to treat bacteria and modify them for a wider spectrum of future applications in biotechnology and biomedical fields, by making them harmless or inserting useful traits into them. The Pro-AG system could be used to engineer a large plasmid; to edit bacterial chromosomal genes; or to insert new cargoes with desired functions along with the gRNA cassette. This could be a powerful new addition to the tools now available for genetic engineering of bacteria.
Journal reference:
Valderrama, J.A., Kulkarni, S.S., Nizet, V. et al. A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus. Nat Commun 10, 5726 (2019) doi:10.1038/s41467-019-13649-6, https://www.nature.com/articles/s41467-019-13649-6