The scientists used as baits certain to pick up the proteins they were in search of from within the human cells in culture in their laboratory. Their catch using 56 types of baits was an amazing 9,000-plus proteins that attach to Rho family proteins.
The Rho proteins were discovered in the 1990s and became famous in being the managers of the cytoskeletal building process. The 20 human Rho proteins are GTPases, enzymes that operate by splitting the energy-storing molecule GTP. They are found all over the inner aspect of the cell membranes. When activated by a signal from outside or inside the cell, they come on to trigger other protein cascades, that in turn stimulate the remodeling of the cell skeleton, by either adding or removing bits according to the framework desired.
Three Rho proteins have been uncovered in detail so far. These are the Cdc42, Rac1 and RhoA. The first is the protein that determines the path of blood cells migrating towards an infected locus. Rac1 is the protein that triggers the cellular apparatus responsible for pushing a cell (other than a muscle cell) forward. RhoA activates the process of forming fibers that aggregate to either pull in a cell’s walls or cause tissues to form a resistant mass, as for instance the fibers that form a blood vessel’s wall.
There are many other proteins in this family, however, and they link up with thousands of others to complete various processes. It was formerly thought that other proteins which bore close similarity to the three listed above would share their function. However, this was later found to be not the case. Instead, variations in the C-terminal ends caused by signal-dependent modifications, and in the cellular organelles they target, cause the biological activity to be quite different from the non-modified forms. It was to explore this area that the current study was begun.
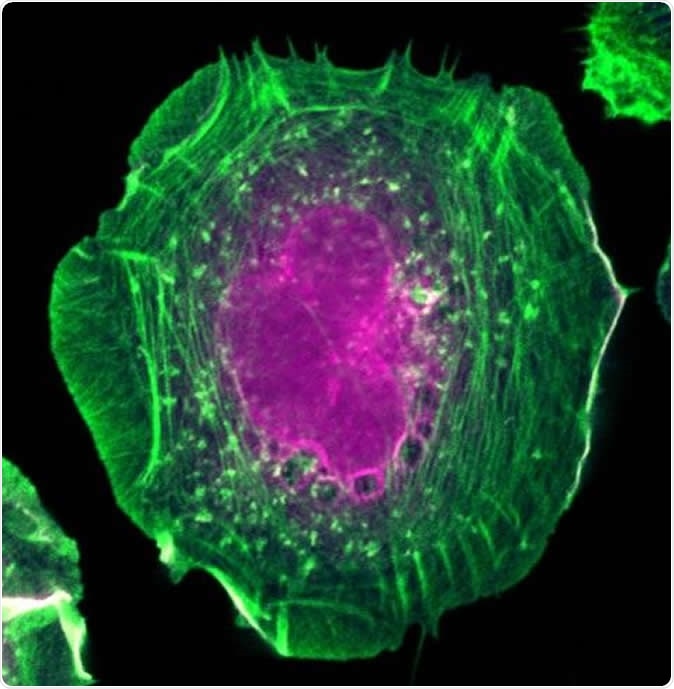
HeLa cell expressing the proximity interaction probe BirA*-Flag-active RAC1. Actin filaments are in green and biotinylated proteins are in magenta. Image Credit: Amélie Robert (IRCM)
The study
The scientists focused on identifying the molecules that interacted with the Rho family of proteins in a proximity interaction network. The Rho proteins are regulated by guanine nucleotide exchange factors (RhoGEFs) and GTPase-activating proteins (RhoGAPs), that switch them on and off as required to bind to the effector molecules.
The scientists used molecules that forced cells in culture to form two-headed proteins. One head consisted of one side of a Rho protein while the other was a biotin ligase enzyme, which attracts other proteins in the vicinity to label them with a biotin moiety using the Rho protein. This process is called proximity-dependent biotinylation. There were 28 of these two-headed proteins.
They also used GTPase enzymes in both active and inactive switch positions. These molecules regulate a host of cellular pathways. The combination of the bait proteins and the GTPases in different configurations enabled the researchers to snag well over 9,000 proteins.
The labeled proteins were then identified singly. To do this, the cells were torn up to release the proteins for analysis. While some of the proteins had already been characterized, such as those which switch the GTPases on and off, there were a multiplicity of other proteins that remained to be defined.
The findings
Among those discovered was the protein that connected different parts of the Rho-dependent cytoskeletal assembly first noticed in the 1990s. The RhoA protein was known to cause another protein called ERM to become activated, causing a type of phosphorylation which in turn made the cytoskeleton more stable. But nobody knew how the ERM protein was activated via RhoA interaction. The current work showed that a protein called SLK connects the RhoA to the ERM protein.
Other unfamiliar proteins dredged up in the current study include GARRE and PLEKHG3. These were found to bind to active Rac1 and RhoG respectively. However, the researchers don’t know exactly how these complexes function.
To find out more about them, the current study described the other molecules they found during their work – so that other laboratories can characterize them and help to reveal their role.
Conclusion
The scientists have not only shown how RhoA-ERM signaling works but demonstrated a unique but successful way to catch a whole lot of proteins. The next step for them is to use this technique to help study how Ras family protein switches function. The Ras superfamily of GTPases, which lies at the heart of multiple cancer pathways is the protein group of which Rho GTPases form a subfamily.
Journal reference:
Bagci, H., Sriskandarajah, N., Robert, A. et al. Mapping the proximity interaction network of the Rho-family GTPases reveals signalling pathways and regulatory mechanisms. Nat Cell Biol (2019) doi:10.1038/s41556-019-0438-7, https://www.nature.com/articles/s41556-019-0438-7