Autistic people are often brilliant, but they may also have poor cognitive skills. The relationship between autism and either condition, inside the brain, has been far from clear. However, now a new study shows that poor cognition in autism could be due to the influence of a small gene fragment.

Image Credit: Nazarova Mariia / Shutterstock
Autism
Autism is widely known for its impact on social and communication skills and is the result of improper nerve connections built during the development of the brain. However, the mental capacity of the affected person depends on their genes. On the other hand, most cases of autism occur as a sporadic occurrence and the cause of the defective gene is unknown.
Says researcher Thomas Gonatopoulos-Pournatzis, “It's very important to understand the mechanisms that underlie autism, especially in idiopathic forms where it is not clear what the underlying causes are.”
The microexon
In previous work, the researchers found a link between short gene segments and autism. These tiny segments, called microexons, were found to be active mostly in the brain. When the gene is transcribed, or copied into an RNA molecule, these microexons are deliberately left out or inserted at the last step by a process known as alternative splicing. These tiny bits of data have a huge impact on how the protein created from the RNA, by a process called translation, is able to bind other complementary molecules during the process of neural development. Yet nobody knows how microexons actually play a part in producing autism.
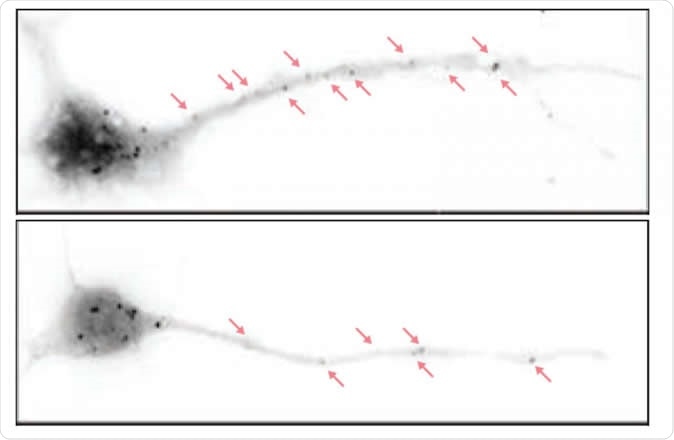
Depicted are hippocampal neurons from a normal mouse (above) and a mouse bred to lack the eIF4G microexon (below). In the latter, there are fewer particles representing paused protein synthesis machineries. In these mice, higher levels of protein synthesis in neurons lead to disrupted brain waves and autistic behaviors as well as cognitive deficits down the line. Image Credit: Thomas Gonatopoulos-Pournatzis
The study
The current study looked at just one microexon which is found in the elF4G gene that drives protein synthesis inside the cell. The researchers analyzed the distribution of this microexon and found that almost all the transcripts of the elF4G gene in the brains of autistic people lacked this microexon.
But they were not done with this discovery. The next step was to find out if this played any role in brain function. To determine this, the team bred mice without this microexon in the brain. They found that without the elF4G microexon, the mice lacked the ability to interact normally with other mice. This being a hallmark of autism, the scientists viewed this as proof that this microexon is required for autism-like brain development.
But the experiment wasn’t done yet. As the scientists continued to test the mice, they found, rather to their astonishment, that these mice also did badly on learning and memory tests. These tests show how well the mice learn to link a given set of surroundings with a stimulus experienced in those surroundings.
Says Gonatopoulos-Pournatzis,” we could not have imagined that a single microexon would have such an important impact not only on social behaviour but also on learning and memory.”
What the microexon does
Next, they delved into the actual structure of the microexon. They discovered that it actually codes for the part of the elF4G gene which permits the gene to associate with the Fragile X mental retardation protein, or FMRP. This is a protein that is not synthesized in people with the Fragile X syndrome, a type of intellectual disability. It is already known that features of autism are seen in about a third of individuals with this syndrome, but the reason for this linkage has only now been discovered.
The explanation is that both elF4G and FMRP are part of a complex that inhibits the binding of further amino acids to form more protein. In this system, a new experience triggers brain activity which in turn remove the brake, according to the researchers. That is, these genes regulate the way the brain responds to a new experience, the system that underlies the fundamental process of learning.
The scientists explain, “This brake in protein synthesis is removed upon experience and we think it allows formation of new memories.” For this brake to function normally, the microexon is an essential if tiny component. Without it, proteins are produced in large amounts, providing plenty of ion channels and protein receptors, as well as the many other signaling molecules that are key to synapse structure and function.
Excessive production of these proteins, as happens when the microexons are absent, blocks certain types of brain waves involved in helping the brain process and understand familiar things to form memory. These findings come from recording the electrical activity of brain slices from mice.
Implications
Confirmation comes from the fact that similar types of proteins are found in higher than normal levels when FMRP is absent. This could mean that both Fragile X and idiopathic autism spring from a common molecular root.
The scientists are understandably excited because this work could explain, for the first time, what causes a large number of autism cases that have hitherto been of unknown cause. Knowing what went wrong is the first step, in many cases, to finding how to fix it. For instance, a novel treatment approach such as small molecules which boost elfF4G microexon splicing in autistic individuals showing this particular defect could well lead to better social and cognitive skills.
Journal reference:
Autism-Misregulated eIF4G Microexons Control Synaptic Translation and Higher Order Cognitive Functions Gonatopoulos-Pournatzis, Thomas et al. Molecular Cell, https://www.cell.com/molecular-cell/fulltext/S1097-2765(20)30006-X