The human body is comprised of billions of cells. Yet it functions and grows as a single organism, because of trillions of interactions that help various cells recognize and respond to each other via different molecules on their surface or inside the cells. Now, a new study published in the journal Nature in April 2020 describes the first-ever interactome map, to help distinguish healthy interactions and disease-producing ones.
The human genome consists of all the genes that function in a human being, normal and abnormal. These genes are transcribed into messenger RNA (mRNA), which carries the genetic instructions for protein synthesis to the cell. The sum of all the mRNA in the cell is called the transcriptome. The transcribed RNA is responsible for protein synthesis, and the sum of all the protein translated from the genome is called the proteome.
However, proteins interact with each other to make the body function, whether in health or disease. The human genome project has provided a list of parts, so to speak, for the cell, but it hasn't explained how these parts fit into the whole equation.
This is the motivation behind the current project – to create a reference map of all the ways that proteins act on each other to build and operate the body. This is the interactome.
Says researcher Marc Vidal, "Since the mid-1990s, our collaborative team has pushed the idea that interactome maps can illuminate fundamental aspects of life." The current paper puts down the first-ever interactome map on which people can build, adding on information as and when it comes in. In other words, the map, as described, is the scaffold on which new data can be added.
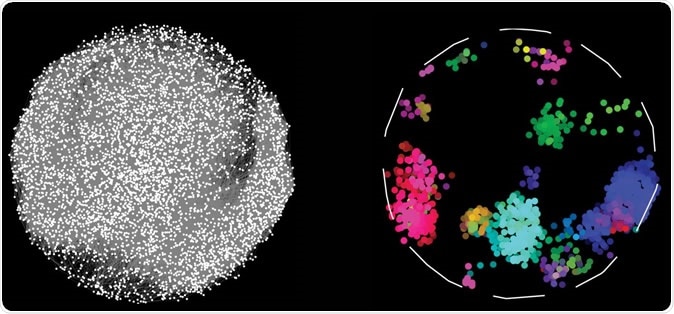
By charting pairwise interactions between 17,500 human proteins, scientists have created a map, on the left, depicting which proteins work together to sustain cellular function. Proteins with similar interactions profiles fall into discrete colour-coded clusters representing different bioprocesses in the cell. Image Credit: Katja Luck et al. / Shutterstock
How did they build the interactome?
The detailed map has taken almost ten years to fill in, and is the fruit of the devoted labors of a large team, comprising over 80 researchers from several different countries. These include Belgium, Spain, France, and Israel besides the US and Canada. Called the Human Reference Interactome (HuRI) map, it details almost 52,570 ways in which 8,275 human proteins interact with each other. This makes it the most extensive such map.
Some of the researchers in the current team had already done part of the work, depicting about 14,000 interactions between different proteins. The current project took this forward, examining the way almost all the proteins that were encoded by human genes interact. This has made the map at least four times as comprehensive.
They first stimulated the simultaneous expression of both genes, encoding a pair of human proteins within a yeast cell. Such pairs were created to express the interactions between almost every human protein if it occurred. Each such interaction gives rise to a molecular switch that enhances the growth of the cell. Thus, yeast cell growth signaled the presence of an interaction.
Using this as a basis, the researchers looked at all the possible combinations of pairs among 17,500 human proteins, looking for interactions between the proteins in each pair. They tested for interactions in 3 distinct versions of a yeast cell, and each test was repeated three times simultaneously. This amounts to testing 3 billion possible interactions, in all.
In the end, they were left with about 53,000 binary interactions between over 8,000 proteins. Most of these were novel interactions. All of them were then validated using another assay.
Why is the interactome important?
The interactome provides a framework to understand how protein molecules interact, causing a range of diseases such as cancer, viral illnesses, and other conditions. The availability of this basic framework helps reveal the fundamental ways in which abnormal gene structure or function disrupts normal cell function to produce the clinical features of disease. For instance, it could help scientists understand how the SARS-CoV-2 virus (which is the agent of the current COVID-19 pandemic) interacts with the proteins of the host cells to produce the manifestations of the disease.
The advantages of this approach are, according to researcher Mike Calderwood, "Genome sequencing can identify the variants carried by an individual that make them susceptible to disease, but it doesn't reveal how the disease is caused. Changes in the interactions of a protein are one possible mechanism of disease, and this map provides a starting point to study the impact of disease-associated variants on protein-protein interactions."
There are altogether about 20,000 genes that code for proteins, but little is clear as of now about the proteins that they encode. However, scientists are using a "guilt by association" principle, in which they consider that two proteins are likely to take part in similar biological processes if they interact with similar proteins. Thus, even the function of an unknown protein can be predicted using the interactome map.
Another researcher, Frederick P. Roth, says humorously, "People can look up their favorite protein and get clues about its function from the proteins it interacts with."
The limitations
The HuRI is the most complete interactome today, but still represents only about 2% to 11% of all the interactions between human proteins. The reason why the majority of interactions are missed is because of the lack of specific molecules that are essential for proper protein function and are found only in human cells.
Even so, HuRI has multiplied the knowledge on protein-protein interactions in human cells threefold or more. This makes it an invaluable source of knowledge in this area.
In fact, scientists are finding that even familiar proteins are being found to have new roles within the cell, and they are discovering molecular-level changes that lead to disease. In this way, HuRI has been fruitful in uncovering hitherto unknown functions for proteins that take part in apoptosis or programmed cell death, the release of cell cargo, and many other cell processes.
Similarly, the team of researchers has attempted to link the data on the various interactions that take place between proteins to the variable expression of genes in each kind of tissue. This has helped them to find the array of interacting proteins that are essential in the development and maintenance of different tissues. It has also helped them identify novel targets for the treatment of a range of diverse disease conditions caused by genetic factors. These could include several types of cancers, as well as certain infectious conditions.
In still another direction, they compared the interactions between various proteins found in cells under study with those found in the reference map, to find out how abnormal proteins found in different diseases triggered abnormal interactions, in turn. This showed up the things that were going wrong to cause that specific condition at a molecular level.
The online portal, on the open-source online publishing site BioRxiv, has already had 15,000 visits since its inception in April 2019. The researchers say many users have already downloaded the whole dataset. Like the previous paper, covering less than a third of HuRI, which has nonetheless been cited more than 800 times, they expect much heavier use of their data in the future.
Journal reference:
Luck, K., Kim, D., Lambourne, L. et al. A reference map of the human binary protein interactome. Nature (2020). https://doi.org/10.1038/s41586-020-2188-x