Researchers at the Stowers Institute for Medical Research have fine-tuned a method to pinpoint surfaces within large multi-protein complexes that are close to, and likely to be directly interacting with, one another.
In a paper published online April 14, 2020, in Cell Reports, members of the Washburn Lab describe the combination of three approaches -- affinity tag protein purification, chemical crosslinking with high-resolution mass spectrometry, and computational molecular modeling with protein docking -- to capture information about neighboring surfaces within the Sin3/HDAC protein complex.
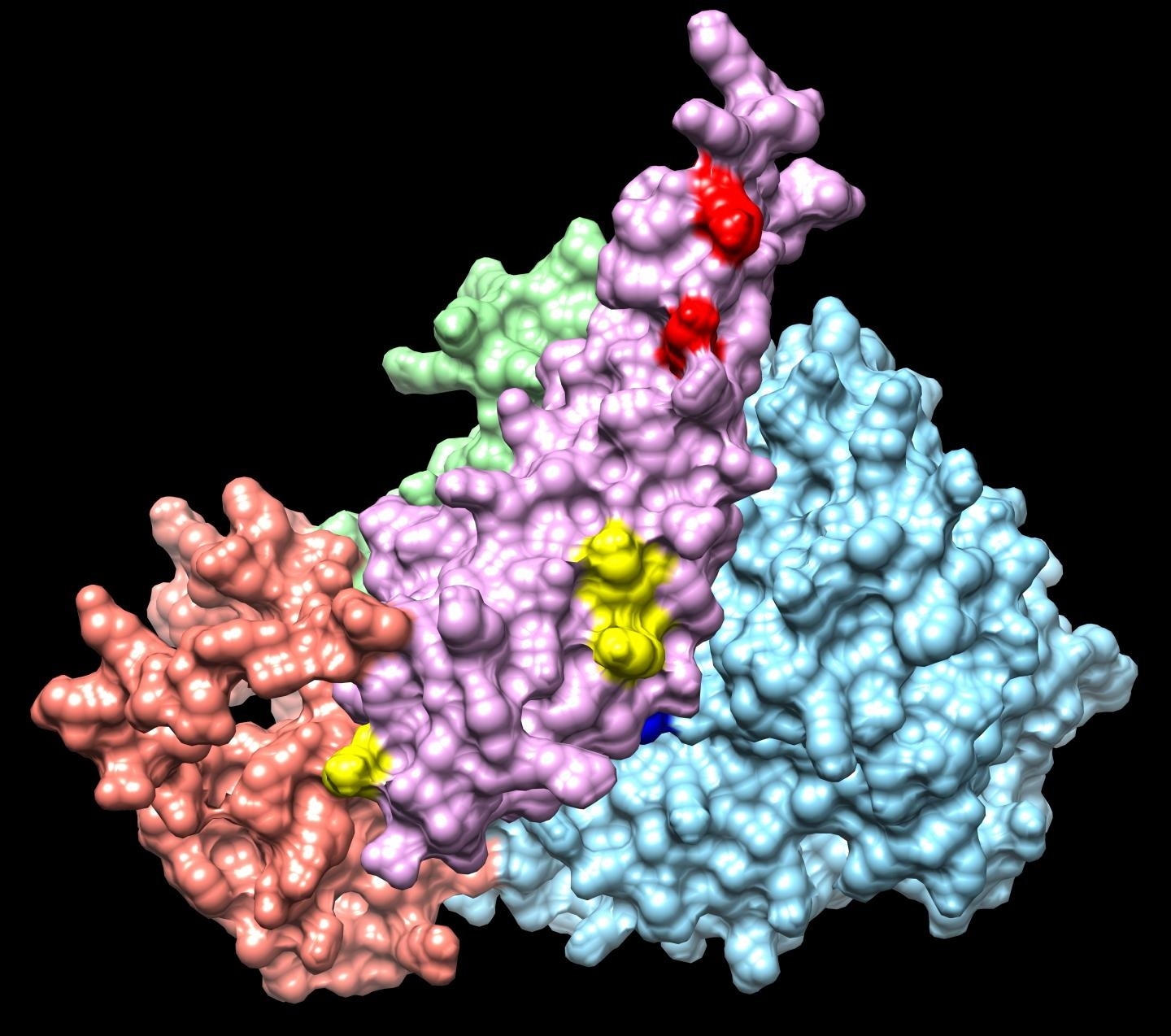 Integrative modeling of a Sin3 complex sub-structure. Image Credits: Washburn Lab
Integrative modeling of a Sin3 complex sub-structure. Image Credits: Washburn Lab
"Putting all these pieces together gives us a new perspective on how protein complexes are put together," with the potential to provide more information more quickly, explains Michael Washburn, PhD, director of the Stowers Proteomics Center.
"The capabilities have all existed, and have been used together a bit, but not in great numbers yet," Washburn says. "It's certainly complementary to existing techniques such as nuclear magnetic resonance (NMR), cryo-electron microscopy (EM), and X-ray crystallography, to really understand how protein complexes assemble, and to see -- how are they really interacting? What happens when you perturb them with drugs or mutations?"
The Sin3/HDAC complex functions in chromatin remodeling and human cancer biology by bringing histone deacetylases (HDACs) to specific locations in the genome and removing acetyl groups from histone tails, thereby suppressing gene transcription. Although the components of the Sin3/HDAC complex have been well defined, high-resolution descriptions of the large multi-protein complex have been difficult to achieve using any one technology.
The problem with mass spectrometry alone, says Charles Banks, PhD, a research specialist in the Washburn Lab, and co-first author of the paper, "is that we can only identify the proteins in the tube. We don't know anything about which proteins are physically interacting with which other proteins."
While assays for individual protein interactions can be done, they are often time-intensive, Banks explains. "What would be really good is if we could capture some sort of structural information about the intact complex before we separate the proteins, and that's where crosslinking comes in."
The crosslinker used in this study, disuccinimidyl sulfoxide (DSSO), can crosslink amino acids - specifically lysine residues - in protein complexes that are within about 30 angstroms of one another. And, says Washburn, it "does something inside the new mass spectrometers that is very clever" - it can be cleaved, essentially providing fingerprints of crosslinked peptides.
"If I see a crosslink between two lysine residues, that means in the fully intact folded protein, those lysines are quite close together, giving us positional information," explains Banks. "And if I've got two different proteins, and two lysine residues - one from each protein - crosslinked together, that means those lysines in those two proteins are usually very close together, which is consistent with the proteins interacting with each other."
Then, says Banks, they are able to use inter-protein crosslink information to try to dock existing crystal structures together, similar to assembling pieces of a 3D puzzle.
In this study, led by Banks and co-first author Ying Zhang, PhD, a scientist in the Proteomics Center, the Stowers researchers isolated Sin3/HDAC complexes from HEK293T cells, using HaloTag affinity purification and the SAP30L subunit as bait. After chemical crosslinking, they found 66 inter-protein crosslinks, and 63 self-crosslinks for 13 Sin3 subunits, of which they were able to use molecular modeling to determine the relative positions of 5 subunits (SAP30L, HDAC1, SUDS3, HDAC2, and ING1) around the Sin3A scaffold.
We think of Sin3A as a platform on which all the other subunits assemble. We still aren't sure which subunits can bind to Sin3A at the same time, although we know that some of them certainly don't bind at the same time."
Charles Banks, PhD, Research Specialist in the Washburn Lab
"But why are there all these other subunits other than the HDACs? Why can't you just recruit the HDACs?" he asks. "We think that probably, the Sin3 complexes control transcription of a specific set of genes, maybe at a specific time and place in specific cells, and we think the other subunits are probably governing that specificity."
When asked about future research directions, Banks replies, "we're a bit limited with our crosslinker at the moment, because it will only capture information based on two lysines being close to each other. It would be really nice to have different crosslinkers that could crosslink between other amino acids, especially if I have another favorite protein that I'd like to study, that does not have many lysines in its sequence."
Molecular modeling with crosslinking information is "like laying the foundation for getting the complete structure of really large complexes, which is hard to do," says Washburn.
"In the future, we can imagine taking what we've done so far and trying to do cryo-EM on those components, and seeing how the two datasets complement each other. Cryo-EM is an amazing technology, and coupling that with crosslinking mass spectrometry can give a much clearer picture of what these molecules look like at the actual interfaces," explains Washburn.
"This is a convergence of technologies that allows us to, going forward, do this on really any protein complex under almost any circumstance"-- even, yes -- toward understanding how coronavirus proteins interact with human protein complexes.
You just have to be able to purify it. If you can get enough material, you can really study any protein complex. It's a matter of doing good biochemistry and preparing a good sample. If you can get really spectacular data, you can build models like this."
Michael Washburn, Director of the Stowers Proteomics Center
"I haven't been this excited about new capabilities in a long time," he says.
Other coauthors of this work include Sayem Miah, PhD, Yan Hao, Mark K. Adams, PhD, Zhihui Wen, Janet L. Thornton, and Laurence Florens, PhD.
This work was funded by the Stowers Institute for Medical Research and by the National Institute of General Medical Sciences of the National Institutes of Health (award RO1GM112639 to MPW and award F32GM122215 to MKA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Lay Summary of Findings
Sin3/HDAC is a large multi-protein complex that is normally responsible for "turning off" a specific set of genes. When its main scaffold is mutated, the complex is also implicated in cancer biology, and is targeted for treatment of many human cancers, including triple negative breast cancer and pancreatic cancer. Although Sin3/HDAC has been well studied and its component subunits have been identified, getting a clear picture of how the subunits assemble has been difficult.
Michael Washburn, PhD, of the Stowers Institute for Medical Research, and his lab have long used proteomics and protein mass spectrometry to study complexes in protein networks. Over the last few years, several technologies have improved, and when combined, allow researchers to obtain unambiguous information regarding the spatial arrangement of neighboring surfaces within multi-protein complexes.
A report from the Washburn Lab, published online April 14, 2020, in Cell Reports, describes the use of three existing approaches -- affinity tag protein purification, chemical crosslinking with high resolution mass spectrometry, and computational molecular modeling with protein docking -- to pinpoint specific surfaces, within the intact protein complex, that are in very close proximity with each other.
In total, they identified 66 inter-protein crosslinks (between two different proteins) and 63 self-crosslinks (within the same protein) for 13 Sin3/HDAC subunits, some of which were mutually exclusive.
The combination of these approaches has been used occasionally in the past, but moving forward, the combined technologies will likely be used much more, allowing researchers to pinpoint interacting surfaces in virtually any protein complex, obtained from any source. These capabilities can be further combined with other powerful techniques such as cryo-electron microscopy to provide an even higher resolution picture of the interactions at interfacing surfaces within intact protein complexes.