Genetic variation analysis and the evidence of recombination events point towards the conclusion that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) might have circulated cryptically among humans for years, as reported by a recent paper on a preprint server bioRxiv.
The pandemic of coronavirus disease (COVID-19) caused by SARS-CoV-2 is an ongoing health crisis and one of the most significant challenges to global health in modern times. This novel coronavirus mainly affects the respiratory tract, potentially causing severe pneumonia and death.
As the virus continues to spread across the world, a myriad of strains have been isolated, and many complete genomes sequenced. A rather small SARS-CoV-2 genome is made up of thirty thousand base pairs. Genomic comparisons with viruses from different patients, animal species, time periods, or places may answer the question we have all been asking ourselves: how did everything start?
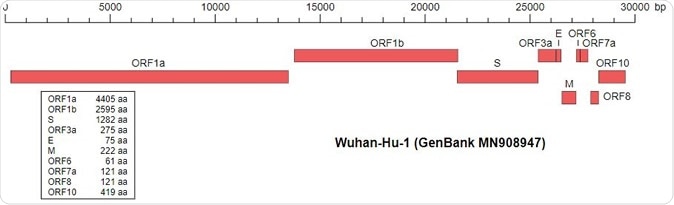
Wu,F., Zhao,S., Yu,B., Chen,Y.-M., Wang,W., Hu,Y., Song,Z.-G., Tao,Z.-W., Tian,J.-H., Pei,Y.-Y., Yuan,M.L., Zhang,Y.-L., Dai,F.-H., Liu,Y., Wang,Q.-M., Zheng,J.-J., Xu,L., Holmes,E.C. and Zhang,Y.-Z. A novel coronavirus associated with a respiratory disease in Wuhan of Hubei province, China. Accession MN908947.
Genomic specificities of SARS-CoV-2
Even though SARS-CoV-2 shares similar genomic traits with other coronaviruses, its sequence is significantly different from some other betacoronaviruses known to infect humans, the novel coronavirus shares 76% amino acid sequence identity with SARS-CoV, 43% identity with MERS-CoV, and 33% identity with HCoV-HKU1.
.jpg)
Novel Coronavirus SARS-CoV-2: This scanning electron microscope image shows SARS-CoV-2 (round gold objects) emerging from the surface of cells cultured in the lab. SARS-CoV-2, also known as 2019-nCoV, is the virus that causes COVID-19. The virus shown was isolated from a patient in the U.S. Credit: NIAID-RML
On the other hand, SARS-CoV-2 shares 96% similarity with a coronavirus collected in Yunnan Province (China) from a bat species Rhinolophus affinis, which is why it is thought that the virus most likely originated from bats. But the mystery remains whether the virus crossed directly from bats to humans, or an intermediate host was implicated in the 'spillover.'

Intermediate Horseshoe Bat (Rhinolophus affinis). Image Credit: Binturong-Tonoscarpe
A recent research paper published in the journal Nature by Dr. Tommy Tsan-Yuk Lam and his colleagues suggested that pangolins should be considered as possible hosts, as they indeed harbor SARS-CoV-2-related coronaviruses. It is rather striking that these pangolin viruses exhibit specific genomic regions very closely related to the human virus – most importantly receptor-binding domain responsible for the attachment and infection of human cells.
Nevertheless, several issues regarding the origin, human introduction point, evolutionary patterns, and driving force of the COVID-19 pandemic remain to be clarified. Recently, a research group from Taiwan tried to tackle these issues in a paper titled 'The origin and underlying driving forces of the SARS-CoV-2 outbreak' that recently appeared on the preprint service bioRxiv, without formal peer review.
A new virus with an ancient pedigree
Dr. Shu-Miaw Chaw from the Biodiversity Research Center (Academia Sinica, Taipei, Taiwan) and her colleagues analyzed 137 genomes of SARS-CoV-2 (as well as related coronaviruses), and found evidence of recent origin and intergenomic recombination. The latter is a process described in a wide array of viruses that may allow specific variants to escape from the current fitness peak and establish a relationship with the new host.
However, the authors state that the sequence similarity of the receptor-binding domain between SARS-CoV-2 and a sequence derived from pangolin coronavirus is probably due to an ancient intergenomic introgression. This means that SARS-CoV-2 might have cryptically been present among humans for years before being noticed recently.
The ancient origin of SARS-CoV-2 is further reinforced by the lack of signatures pointing towards the adaptive evolution, as demonstrated by frequency spectra in samples from the recent outbreak.
"For a recently acquired virus, rapid evolution and a strong signature of positive selection are expected," caution study authors. "For example, during its short epidemic in 2002-2003, several rounds of adaptive changes have been documented in SARS-CoV genome", they add.
Understanding the evolutionary path
Unlike adaptive evolution that was previously shown for SARS-CoV during the brief SARS epidemic, the analysis of SARS-CoV-2 genomes reveals signs of relaxation of selection. After host adaptation, the virus may evolve following such relaxed or purifying selection seen in this novel coronavirus, which is one of the evolutionary driving forces.
Hence, further work is crucial to sequence a large number of samples from the early outbreak event, as well as to examine hospital archives in pursuit of traces of SARS-CoV-2 ancestors.
"This information not only can help us to understand the evolutionary path of this virus but also unravel the critical steps for it to achieve effective spreading in humans," explain study authors. "Nature never runs out of material to create new pathogens. It is not whether but when and where the next epidemic will occur," study authors conclude.
Journal references:
Main source:
Chaw, S.M. (2020). The origin and underlying driving forces of the SARS-CoV-2 outbreak. bioRxiv preprint server. https://doi.org/10.1101/2020.04.12.038554
Additional source:
Lam, T.T. (2020). Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. https://doi.org/10.1038/s41586-020-2169-0