The novel coronavirus that is circulating the world at present is called severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). As the causative agent of COVID-19 disease, it has killed at least 217,000 people out of the 3.11 million reported cases so far.
When the virus enters the host cell, it takes over the cell machinery for its replication. In order to do this, the enzymes encoded by its RNA bring about significant changes to the mRNA molecules being transcribed from its genome. This is by synthesizing a cap over the 5’ end of the mRNA.
Why Is RNA Capping Important?
The formation of an RNA cap in coronavirus only occurs when several nonstructural proteins take part, such as nsp13, helicase, and nsp16. These are all enzymes with different functions, such as phosphatase, helicase, exonuclease, and methyltransferase activity.
With the SARS coronaviruses, nsp16 adds a methyl group to the 5’ end of the mRNA after forming a complex with another nonstructural protein, nsp10. This complex then quickly converts the mRNA from an uncapped to the capped form by transferring a methyl group to the first nucleotide, usually adenosine, on the ribose 2’- O position of the newly forming mRNA strand.
The capped mRNA can now avoid recognition by the innate immune system, the methylation allowing it “to mimic cellular mRNAs.” Logically, preventing this activity should, therefore, restrict the ability of the coronavirus to cause disease by allowing the immune system to recognize and react to the infection.
In fact, vaccination with SARS viruses that lack nsp16, or using antibodies to disrupt the nsp16-nsp10 complex, allows mice to survive what would normally have been a lethal dose. Thus, the complex could be a promising target for vaccine development or therapeutic drugs.
However, the mechanism by which this mRNA capping in coronaviruses is catalyzed is unclear. The current study was aimed at uncovering this process.
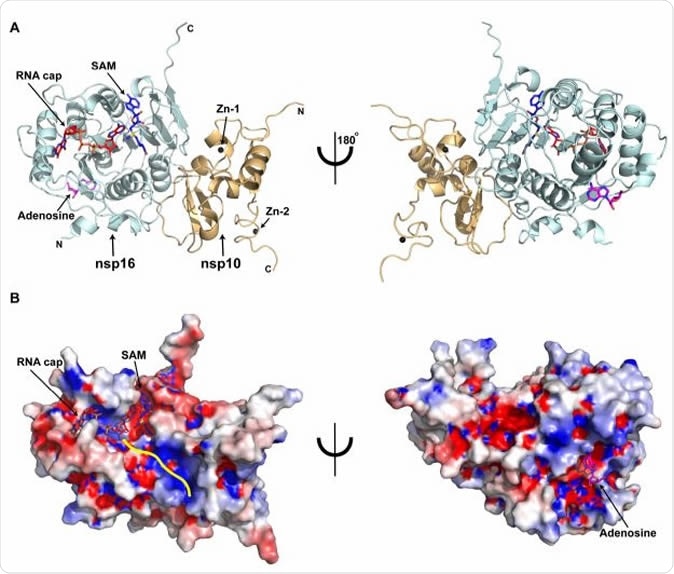
Overall structure of the SARS-CoV-2 nsp16/nsp10 ternary complex A) Subunit arrangement of nsp16 (cyan) and nsp10 (orange cartoons) proteins with respect to RNA cap (red), and SAM (blue). Proteins, nucleotides are shown in cartoon and stick modes, respectively. Black spheres, Zn2+ atoms bound to nsp10; Magenta stick, adenosine bound to nsp16. B) Electrostatic surface representation of nsp16/nsp10 with saturated blue and red at +5 kT/e and -5 kT/e, respectively. A yellow line represents a tentative path for downstream RNA sequence calculated by superposing the target bases in current, VP39, and Dengue NS5 ternary complexes.
How Was the Study Done?
The scientists tried to understand the structure of the nsp16-nsp10 complex formed with the methyl donor S-adenosyl methionine (SAM) and its target, the RNA cap analog.
They also crystallized the complex along with a range of approved drugs to test its binding. These drugs can bind to the SAM binding site because they have the same binding site characteristics, a purine ring with a ribose sugar.
They soaked the crystals with an RNA cap analog to serve as the substrate for this complex.
What Did the Study Reveal?
The researchers determined the high-resolution ternary structure of the full-length nsp16-nsp10 heterodimer complex and found that SAM and the RNA cap were attached in their binding pockets. A potential adenosine binding site was also observed on the other face of nsp16, partly containing unique amino acid residues.
The crystal structures of the nsp16-nsp10 complex suggest a mechanism whereby the methylation of the target adenine of the RNA cap is catalyzed. The SAM molecule has a methyl group near the 2’-OH of the first adenine base, and this methyl group is, therefore, open for direct in-line attack by this hydroxyl group.
The supporting evidence for this hypothesis includes the strong 2’-O methyltransferase activity of the nsp16-nsp10 complex. When the first nucleotide was changed from adenine to guanine, the activity shows a steep drop, as recorded in an earlier study. This ternary structure shows why this decline occurs.
The guanine at this position interferes with the methyl transfer because of the unfavorable re-orientation of the target sugar 2’- OH, as a result of the repulsion between the positively charged sulfur of the SAM and the purine ring of guanine.
The structure had all the components for methylation of the 2-O’ site of the ribose molecule of the first nucleotide of SARS-CoV-2 mRNA, even though this event was not observed. The absence of methylation may reflect the lack of another downstream RNA sequence.
The study also shows important changes occurring in the conformation of this enzymatic complex as the RNA binding site attaches to its substrate, as it shifts from a binary to a ternary state. It also throws light upon the mechanism of methylation of the RNA cap at the 2’-O ribose site.
They showed the presence of mutations across the whole length of the nsp16 sequence. Three of these mutations are located in the adenosine binding pocket, two of which are unique to the SARS-CoV-2.
One of the drugs examined, called adenosine, binds to this allosteric site that is unique to the nsp16-nsp10 complex of the SARS-CoV-2. As such, this kind of complex could become the basis for developing new therapeutics from nucleoside analogs. This site may be an alternative site for the development of antiviral therapies.
Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.