With the novel coronavirus continuing to take a shocking toll of lives in the developed as well as in the developing world, scientists are racing to find newer, better ways to identify those affected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) both symptomatic and asymptomatic, as well as to treat patients with potentially severe infections.
Currently, the gold standard test for viral detection is the quantitative reverse transcription-polymerase chain reaction (RT-qPCR) assay. This technology is the most accurate among various testing approaches. However, as currently performed, it has the serious drawback of involving the use of high-end equipment used to isolate nucleic acid and real-time PCR thermal cyclers. These put it out of reach in the very regions which need it the most because of high transmission rates.
Many low- and middle-income countries are not equipped with the resources for this type of rapid and reliable testing, allowing many cases to go undiagnosed, increasing the likelihood of high community transmission rates.
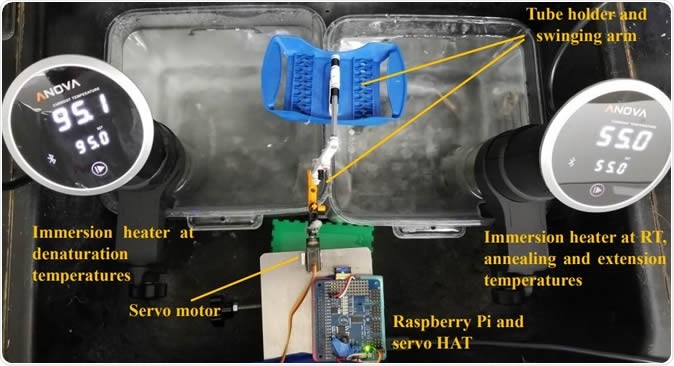
Water bath setup for SARS-CoV-2 detection using RT-PCR. Sous vide immersion heaters provided sufficient and consistent temperatures for RT, denaturation and annealing/extension steps. A Raspberry Pi controlled a servo motor that moved the PCR tubes between the baths with a cell phone app.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Using Inexpensive Technology For Accurate Results
The current study is aimed at introducing an innovative substitution for the old method of transferring the reaction tubes through a series of water baths. Instead, the researchers used just two baths, which were converted plastic containers meant for food storage, fitted with sous vide immersion heaters.
These heaters can heat water to 99 degrees Celsius, which denatures nucleic acid and amplicons.
The advantage of this setup is the ease and convenience of use, coupled with the capacity to adjust the temperature accurately while maintaining heat circulation. The researchers used a Raspberry-Pi to devise a means of controlling a motorized arm that shuttles the reaction tubes between the baths.
The difference in cost is noticeable: the current device costs a matter of a few hundred dollars, in contrast to the thousands of dollars required for conventional PCR thermal cyclers.
Increasing Speed Without Sacrificing Reliability
There are other apparent advantages: the current device can handle a maximum of 96 samples in a single step, and does so very fast, completing 40 cycles based on the TaqMan probe within half an hour. It uses the available polypropylene PCR tubes for this purpose.
If the laboratory desires, thin-film PCR reaction tubes such as Cepheid SmartCycler or glass capillary tubes may be used to cut the time down further to just 12 minutes. This comprises 10 minutes for the PCR with just 2 minutes for the RT.
To test this approach, the investigators used samples that were inactivated by heat or untreated, directly adding them to single-step RT-PCR master mixes, which do not require the extraction of RNA. In addition, they came up with a rapid extraction step, which requires only 3 minutes, using magnetic particles, to generate RNA templates.
What Were the Study Results?
The performance so far outclasses most benchtop thermal cyclers. First, the experiment confirmed the ability of sous-vide heaters to maintain the water bath temperature at 60 °C and 97 °C, for 30 minutes or more, with only 0.1 °C and 2 °C variation respectively. Appropriate time and temperature settings would be required at high altitudes.
The relative viral load of each sample was predetermined using commercial RT-PCR systems. The reaction time was 1 hour 22 minutes. Untreated media samples and extracted RNA samples produced matching qualitative PCR results.
Using COVID-19 positive clinical specimens, the scientists showed that within just 12 minutes, the samples showed increased fluorescence, when thin-walled tubes were used, and within 26 minutes with regular PCR tubes.
When tested with and without prior heat inactivation at 95 °C for 10 minutes but skipping the RNA extraction step, the former was found to produce excellent results.
Finally, they used a rapid (3 minute) RNA extraction step using a magnetic particle-based technique. This showed a 100% sensitivity in a 12-minute run.
What Do the Findings Imply For COVID-19 Management?
The researchers say that their data suggest the use of a water bath-based RT-PCR technique to allow the completion of the reaction in 12 minutes. When a 3 minute RNA extraction is done, it allows 100% sensitivity: with raw heat-treated samples, 90%; and with untreated samples, only 70%. An RNA extraction step will heighten the yield with low viral loads or untreated samples.
The potential disadvantage is the consumption of larger volumes of certain reagents, and a more extended period if regular PCR tubes are used. Nonetheless, the researchers summarize: “This would allow locations with no access to real-time PCR thermal cyclers or even basic thermal cyclers to perform highly sensitive and specific gold-standard RT-PCR assays for COVID-19.” This could close the loop and allow developing countries to boost their rates of testing, thereby identifying the population to be isolated to prevent massive rates of viral spread which could potentially overwhelm their low-capacity healthcare systems and increase the number of unnecessary deaths.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources