The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is thought to have originated from an animal species, most likely bats, which succeeded in crossing over to humans. However, the exact species of origin has not yet been confirmed.
The genome of the SARS-CoV-2 virus is about 30 kB, with over two-thirds composed of genes for nonstructural proteins. The remaining third encodes the spike, envelope, membrane, or nucleocapsid proteins.
The virus sequence was first generated within a few weeks from the first known case, and in a little over four months, more than 16,000 sequences have been uploaded to universal data banks.
Understanding the Mutations in Viral Proteins
The present study aimed to determine the highly prevalent amino acid substitutions in viral proteins from the available sequences of SARS-CoV-2 in keeping with the known timeline of the pandemic. These were evaluated with respect to their significance for viral fitness.
The SARS-CoV-2 sequences for the study came from the GISAID database and included the 12,562 high-quality complete sequences available on May 3rd, 2020, arranged in chronological order of isolation. The researchers also used one reference sequence each from SARS-CoV, pangolin, and civet, and three from bat coronaviruses, using Genbank.
The next step was to analyze the non-synonymous mutations and select those suitable for further study. The researchers arbitrarily set the day when the first amino acid substitution was reported at the end of February 2020 as representative of an early date for the pandemic. They also arbitrarily chose a cut-off frequency of 10% to determine whether an amino acid change was to be called widespread.
Having set these limits, they selected any substitution, which was “widespread” after this “early date” for study. They also noted the continent-based distribution of the identified variants to study their geographical location.
Each substitution was analyzed by aligning it with the corresponding proteins of the following coronaviruses: bat, pangolin, civet, and SARS, to identify the location of the change, whether in the conserved or variable regions.
What Did the Researchers Find?
The study showed eight amino acid substitutions across the viral genome, which appeared before the end of the early phase of the pandemic. These spread out to be found in 10% of known isolates.
Seven of these were in structural proteins and one in a nonstructural one. Four were present in January in China, the others in Europe by the second half of February but in other continents by the end of the next week.
One mutation was found in nearly all the samples, namely, Asp614Gly in the spike protein. However, all mutations were found in all continents except for 175Met, which was not found in Africa.
Most of the mutations were found at sites that were conserved in both SARS and similar coronaviruses.
Compared with the major amino acid changes in early, mid, and late phase-sequences of the SARS epidemic, the researchers found that the non-synonymous mutations that became common in the majority of SARS-CoV appeared different. Among 11 substitutions that became widespread, almost all of them were in nonstructural proteins, but three affected the spike protein. While they were found in conserved positions of the genomes of the bat and civet CoVs, this was not the case with the pangolin CoV.
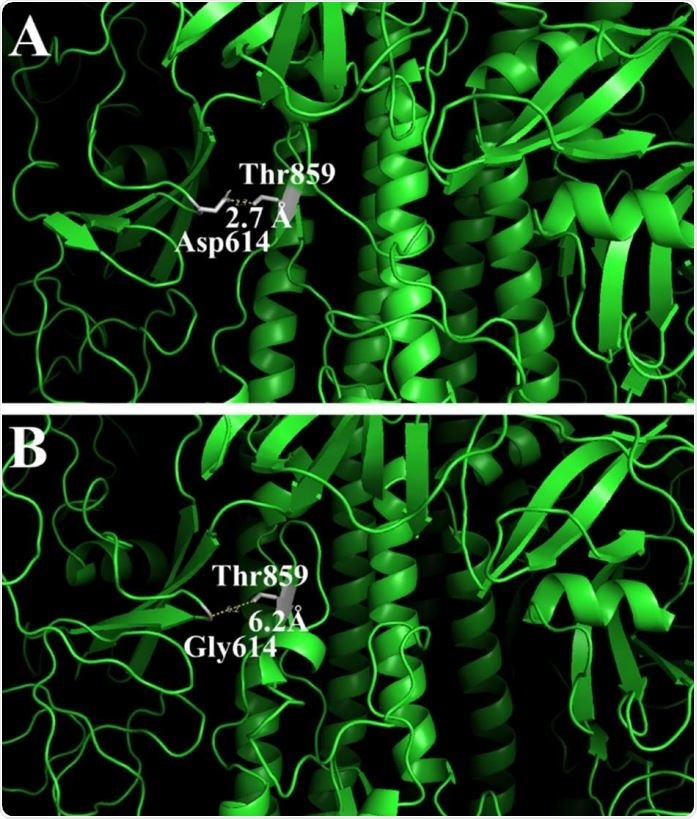
Changes in the 3-D structure of the S-protein in the original (A) and mutant (B) proteins. The pictures show amino acid residues on 20Å distance from Asp614 (A) or Gly614 (B) and their distance to Thr859.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
How Did the Mutations Affect Protein Structure or Function?
The researchers constructed models for both the original and mutant spike proteins, assessed their accuracy, and looked for changes in the physical structure as well as interatomic distances to study various aspects of the protein. For instance, if the mutation affected phosphorylation sites or motifs, this could help them decide on its neutrality or harmfulness.
Alterations in the secondary structure of the protein were also predicted. If the mutation was in or near a site known to serve as an antigen, the impact on the antigen was assessed as well using appropriate prediction software.
The researchers also found that the Asp614Gly mutation became almost universally prevalent by the end of April 2020. This mutation is found in the S1 domain of the S or spike protein and results in a more relaxed structure with a cavity, but without any difference in the antigenic potential of the epitope.
The N or nucleocapsid protein showed a double substitution, which became more prominent during the pandemic, which was predicted not to have any significant effect, however.
In the M or matrix protein, the mutation consisted of the substitution of Thr175 by Met175. This was predicted to be a potential phosphorylation site, with the mutation being predicted to be harmful. This was present along with the 203KR204 mutation in 98% of the cases. The rapid fade-out of this mutation supports the reduced viral fitness associated with it.
Other mutations were also predicted to be neutral.
Commonly Found Mutations In SARS-Cov-2 Affect Conserved Sites
The researchers consider the current pandemic to be an example of the worst-case scenario where a new infectious agent emerges in a population without any immunity, leading to very rapid transmission with insignificant selection pressure from the immune response. Under such circumstances, many different variants of the virus would be expected to arise, with the mutations that increase viral fitness is expected to gain prominence.
The researchers arbitrarily determined cut-off criteria to distinguish sequencing errors and random substitutions from those which could affect viral fitness much more significantly. It carries the inherent risk that some less common mutations will be overlooked.
The study shows that 7 of 8 prominently found mutations occur in highly conserved residues in coronaviruses that are related to each other. This should indicate their high impact on viral replication and survival, but a significant difference from those found in SARS-CoV is that they built up in structural proteins.
The researchers comment, “Interestingly, most of these mutations faded out, except for the Asp614Gly in the S protein that became predominant, suggesting that it contributed to viral fitness. Some others are still increasing in prevalence.” This information may help to track SARS-CoV-2 as it continues to change and spread in different populations.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.