As of now, the virus has caused over 5 million confirmed cases and over 332,000 deaths. In the absence of either an effective vaccine or a therapeutic drug targeting the virus, non-pharmacological interventions (NPI) have been the mainstay of containment measures to prevent the spread of the epidemic.
Identifying Viral Antigenic Sites
The unusually specific nature of antibody binding makes monoclonal antibodies (mAbs) a good choice to attach the exposed antigenic sites of the virus, thus preventing their entry into the host cell and so averting infection. The effectiveness of mAbs directed against viral surface proteins has been shown for SARS, MERS, and Ebola viruses.
With SARS-CoV-2, the spike (S) protein on the viral surface is a significant component of the cell entry mechanism. The S protein has two subunits, S1 and S2. The S1 subunit contains the receptor-binding domain (RBD), which is the attachment site for the ACE2 host cell receptor. The S2 subunit is responsible for the fusion of the virus with the host cell membrane, facilitating viral entry into the host cell. This interaction of the virus with the receptor is the main target for neutralizing antibodies and is, therefore, the primary candidate for the development of therapeutic antibodies.
Using Monoclonal Antibodies to Study Viral Antigens and Prevent Infection
The best therapeutic or vaccine can be designed only if the antigenic site is fully delineated via neutralizing monoclonal antibodies that target all the epitopes of the viral antigen. Such antibodies are seen in the serum of humans while they are infected or recovering and in immunized animals.
A combination of several noncompetitive mAbs may be optimal to achieve effective treatment of an exposed patient, by using a variety of mechanisms to prevent viral infection. This would prevent the emergence of escape variants of the neutralizing antibodies as well.
The current study describes the isolation of a set of neutralizing mAbs against the RBD of the virus, selected from the library built on patient samples acquired in the acute phase of the illness. These are found to specifically bind to different epitopes of the antigenic site, which indicates they could be used for the development of drugs and vaccines.
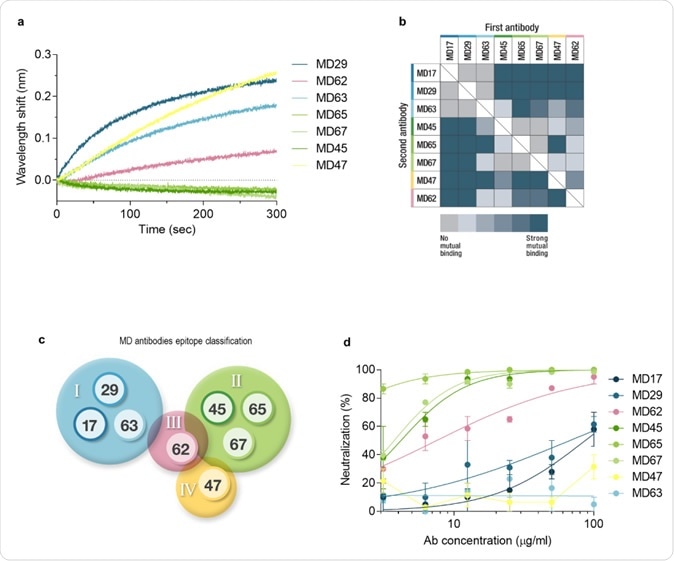
Epitope binning and SARS-CoV-2 neutralization. a. Biolayer interferometry was applied for the epitope binning experiments. Representative assay results are shown for MD65 mAb. The purified antibody was biotinylated, immobilized on a streptavidin sensor, and saturated with RBD. The complex was then incubated for 300 sec with each one of the indicated antibodies. Time 0 represents the binding to the MD65-RBD complex. b. Complete epitope binning of the eight selected MD monoclonal antibodies. Binding was evaluated by the ability of each pair of antibodies to simultaneously bind RBD, using biolayer interferometry. c. Four non-competing RBD binding epitopes were identified and accordingly classified into four groups: I (blue), II (green), III (pink), and IV (yellow). d. SARS-CoV-2 in vitro neutralization using plaque reduction neutralization test (PRNT). Neutralization potency was determined by the ability of each antibody (at indicated concentrations) to reduce plaque formation; results are expressed as percent inhibition of control without Ab.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Finding mAbs Against SARS-CoV-2
The researchers found two samples taken from patients with severe disease, which showed high titers of antibodies that bound to the RBD and neutralized the virus. These were the basis of the antibody phage display library, consisting of about 10 million different antibodies. They then performed panning in three steps to enrich neutralizing antibodies against S1 and RBD. The resulting antibodies were assayed for their ability to bind S1. These antibodies alone were then fully expressed with single-chain Fv and Fc portions.
The final analysis was carried out on eight antibodies with unique sequences that were specifically bound to the RBD and the S1 subunit but not to the N-terminal domain of the S protein. The specific binding affinity for RBD was highest and lowest for the antibodies MD29 and MD67, respectively.
The RBD of the spike protein of the SARS-CoV-2 binds to the virus, but viral infection can be blocked by neutralizing antibodies acting on other epitopes as well. The selected mAbs were classified by the specificity of binding for each epitope.
Each antibody-RBD combination was challenged by all the other antibodies one by one to see which would co-bind to the RBD at an overlapping but distinct epitope. The presence of almost identical or distinct epitopes was discernible by the wavelength shift in the pattern of interference.
This paired-antibody study showed which antibodies bind to the same epitope. This gave rise to four groups based on epitope recognition. These four groups were also classifiable by the similarity observed in their sequences.
The researchers also chose to use the antibody CR3022, which targets an epitope on the RBD to identify further the epitopes recognized by the selected mAbs. Using a recombinant version of CR3022 loaded with RBD, along with the selected mAbs, they identified three antibodies (MD17, MD29, and MD63) which fail to bind to the RBD-linked CR3022 epitope in the presence of CR3022. This indicates that their binding sites fall within this epitope.
The ability of these three antibodies to neutralize the virus was tested, and it was found that only two out of three were neutralizing, though all had homologous sequences. The reason for the missing antibody maybe its reduced affinity for the epitope compared to the other two. They note that CR3022 is also a non-neutralizing antibody for the same reason.
The study has thus resulted in the isolation and further description of the characteristics of a set of human SARS-CoV-2 neutralizing antibodies that bind four different epitopes on the RBD. Neutralizing antibodies are valuable for the treatment of exposed individuals as well as to treat human infections caused by CoVs.
These could, therefore, be developed into efficient preventive or therapeutic agents. The difference in target epitopes also shows the potential to combine these antibodies to enhance treatment and prevent the risk of the emergence of escape mutants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
NOy-Porat, T., et al (2020). Tiger Team: A Panel of Human Neutralizing mAbs Targeting SARS-Cov-2 Spike At Multiple Epitopes. bioRxiv preprint doi: https://doi.org/10.1101/2020.05.20.106609.
- Peer reviewed and published scientific report.
Noy-Porat, Tal, Efi Makdasi, Ron Alcalay, Adva Mechaly, Yinon Levy, Adi Bercovich-Kinori, Ayelet Zauberman, et al. 2020. “A Panel of Human Neutralizing MAbs Targeting SARS-CoV-2 Spike at Multiple Epitopes.” Nature Communications 11 (1). https://doi.org/10.1038/s41467-020-18159-4. https://www.nature.com/articles/s41467-020-18159-4.