The unfolding COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in a global crisis with medical, economic, and humanitarian consequences. The viral agent belongs to the group of betacoronaviruses and causes a (potentially fatal) respiratory disease.
Viruses are predicted to quickly adapt to a new species by a series of mutations that increase transmissibility. However, not much is known about the tempo and mode of evolution at the outset of many outbreaks - including the ongoing COVID-19 pandemic.
.jpg)
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of an apoptotic cell (pink) heavily infected with SARS-COV-2 virus particles (green), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Analyzing mutations to forecast virus behavior
Knowing both the type and the frequency of mutations at the start of a pandemic can provide key information on how exactly a newly introduced pathogen is either well-adapted or mal-adapted - potentially informing treatment options that will make use of its weaknesses, but also avoid the evolution of drug resistance.
Therefore, the set of mutations that we can discern during this pandemic may decipher how the virus will adapt to human hosts as it continues to spread. And due to a relatively high mutation rate pertinent for RNA viruses, comparing genome sequences may give us a wealth of information.
Obtaining such important insights was the primary aim of a small research group from the University of Pittsburgh, Center for Evolutionary Biology and Medicine, and Center for Vaccine Research in Pittsburgh, USA.
A deep dive into sequencing data
In their methodological approach, the researchers mapped SARS-CoV-2 mutations onto the tips of the phylogenetic tree and generated them back until they coalesced at a common ancestor. This enabled them to precisely count the number of independent substitutions inherited by one or more viral strains.
Complete SARS-CoV-2 genomes were downloaded at the start of May 2020 from GISAID, which is a collaboration that provides timely distribution of genetic sequencing data related to COVID-19 and influenza in a freely accessible database.
Viral genomes with more than 500 degeneracies were eliminated, resulting in a collection of 12,435 genomes; of those, 12,285 were with a known date of collection and utilized as a proxy for the time scale of growth relative to the first genome.
Various programming languages and web-based platforms for the analysis of the SARS-CoV-2 genome were used. A fully open-source and reproducible analysis pipeline was provided on the GitHub repository.
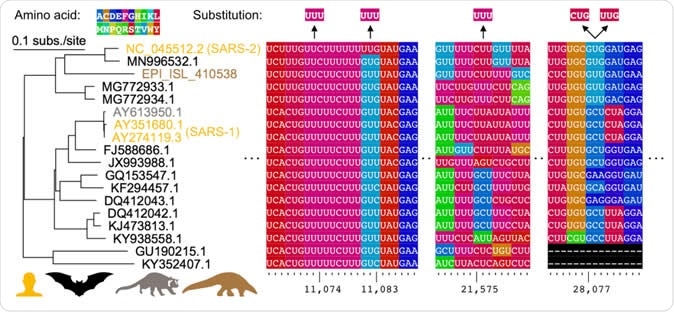
Comparative genomics of parallel substitutions in protein-coding regions. A midpoint rooted maximum likelihood phylogenetic tree constructed from SARS-related coronavirus genome sequences with accession numbers colored by host species (human, bat, masked palm civet, or pangolin). Codons are colored by their corresponding amino acid with frequent nucleotide substitutions shown relative to the reference SARS-CoV-2 sequence (top). Poly(U) sequences are found surrounding the substitutions at positions 11,074, 11,083, and 21,575. The two substitutions observed at position 28,077 results in conversions to the same amino acid in ORF8.
An expose of early human adaptations
"In this study, we determined that SARS-CoV-2 is evolving predominantly under purifying selection that purges most mutations since they are deleterious", explains study authors.
"This suggests that SARS-CoV-2 was well-poised to invade the human population, although it continues to adapt to humans through specific mutations that may accumulate in individual genomes as SARS-CoV-2 continues to evolve", they add.
Still, it has to be noted that several parallel mutations arose in numerous independent lineages, which may provide a substantial fitness advantage over the ancestral genome.
In any case, the small number of mutations altogether suggests that coronaviruses are well versed in jumping between various hosts; therefore, precautions should be taken in order to evade any contact with their known animal reservoirs.
This is further reinforced by the fact that there is only a handful of genome positions where multiple substitutions have been observed, despite an abundant supply of mutations. Such a low mutation rate of SARS-CoV-2 provides us with hope for an eventual vaccine.
"The few highly parallel substitutions that we observed offer intriguing avenues for further investigation, as most are cryptic and located in poorly characterized regions of the SARS-CoV-2 genome", conclude study authors.
The lack of mutations in large portions of the virus may pinpoint potential targets for drug development. And while we all brace for the potential second wave of the COVID-19 pandemic, the insights from this study are of utmost importance for steering further research endeavors.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.