The coronavirus disease (COVID-19) pandemic, caused by the SARS-CoV-2 virus, has resulted in expedited research efforts to develop diagnostic approaches, treatments, and vaccines to mitigate this global health emergency swiftly.
A key target of most of these efforts is the S-protein of the virus – a large trimeric class I fusion protein that is metastable (i.e., sensitive to small disturbances) and somewhat cumbersome to produce recombinantly in large amounts.
Deciphering the S-protein
In a nutshell, S-protein binds to a host-cell receptor and prompts cell entry via fusion of host and viral membranes. The primary SARS-CoV-2 receptor is the angiotensin-converting enzyme 2 (ACE2) ubiquitously expressed in a myriad of human organs and cells.
The S1 subunit of S-protein contains the receptor-binding domain (RBD), which recognizes the receptor. Subsequently, the S2 subunit mediates membrane fusion and has an additional protease cleavage site. After RBD binds to ACE2, S2 rearranges and becomes highly stable.
However, a stabilized conformation of class I fusion proteins is highly desirable for vaccine development, because it is found on infectious virions and displays surface molecules that can be targeted by antibodies to avert the entry process.
Previous research has shown that prefusion stabilization has a tendency to increase the recombinant expression of viral proteins, primarily by blocking the misfolding process that stems from a tendency to take on a more stable postfusion structure.
This is why scientists from the University of Texas at Austin in the United States decided to employ structure-based design in order to increase the stability and yield of the SARS-CoV-2 spike ectodomain in the conformation before the fusion.
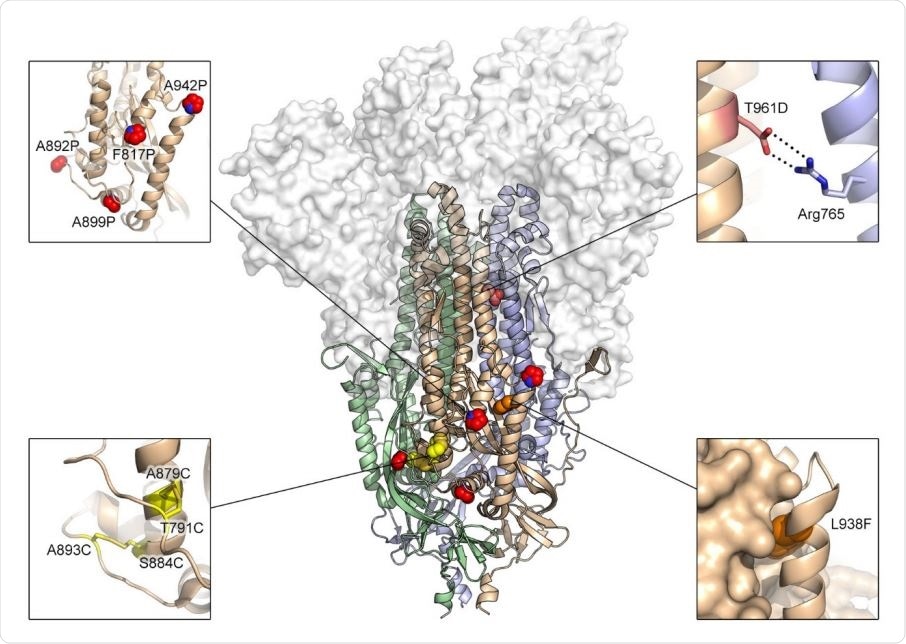
Exemplary substitutions for SARS-CoV-2 spike stabilization. Side view of the trimeric SARS-CoV-2 spike ectodomain in a prefusion conformation (PDB ID: 6VSB). The S1 domains are shown as a transparent molecular surface. The S2 domain for each protomer is shown as a ribbon diagram. Each inset corresponds to one of four types of spike modifications (proline, salt bridge, disulfide, cavity filling). Side chains in each inset are shown as red spheres (proline), yellow sticks (disulfide), red and blue sticks (salt bridge) and orange spheres (cavity filling).
Structure-based design of protein variants
To achieve this goal, this research group designed over one hundred structure-guided S-protein variants, in accordance with the previously determined structure of the prefusion SARS-CoV-2 spike by cryogenic electron microscopy.
They basically focused on engineering the S2 subunit, since it goes through a large-scale refolding to smoothen the membrane fusion. One of the employed strategies was the introduction of disulfide bonds in a region that alters conformation before and after fusion.
Different strategies were employed to enhance protein stability without influencing the overall fold. After that, the feasibility of large-scale production in various cell lines was tried for best-performing modifications.
High stability with preserved antigenicity
Structural, biophysical, and biochemical characterization of protein variants identified manifold individual substitutions that augmented both protein stability and yields.
The best variant was named HexaPro and contained six beneficial proline substitutions, leading to the ten-fold higher expression when compared to its parental construct. Furthermore, it was able to withstand heat stress, multiple freeze-thaw cycles, as well as storage at room temperature.
When the structure of HexaPro was assessed with high-resolution cryogenic electron microscopy, it was confirmed that it retains the prefusion spike conformation. All of these findings disclosed HexaPro spikes as a rather promising tactic among subunit vaccine candidates.
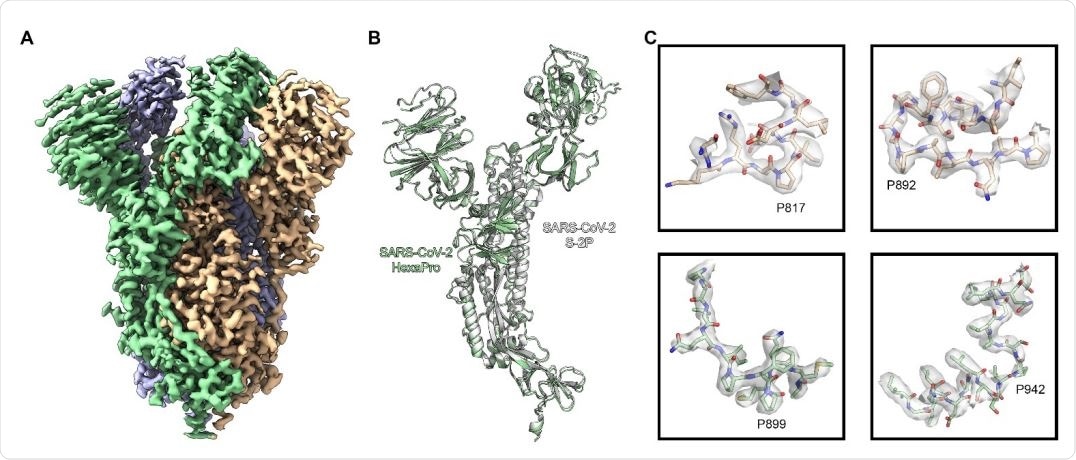
High resolution cryo-EM structure of HexaPro. (A) EM density map of trimeric HexaPro. Each protomer is shown in a different color; the protomer depicted in wheat adopts the RBD-up conformation. (B) Alignment of an RBD-down protomer from HexaPro (green ribbon) with an RBD-down protomer from S-2P (white ribbon, PDB ID: 6VSB). (C) Zoomed view of the four proline substitutions unique to HexaPro. The EM density map is shown as a transparent surface, individual atoms are shown as sticks. Nitrogen atoms are colored blue and oxygen atoms are colored red.
Towards the next-generation vaccine design
"In our HexaPro cryogenic electron microscopy dataset, we observed a third of the particles in a two-RBD-up conformation," explain study authors the main discovery of their bioRxiv preprint paper.
And indeed, this had not been previously observed for SARS-CoV-2 spikes – until a recent structure of a modified spike containing four hydrophobic substitutions was determined, which brought subdomain 1 closer to S2 subunit.
"We hypothesize that the more stable S2 in HexaPro allowed us to capture this relatively unstable conformation that may transiently exist prior to triggering and dissociation of S1", say study authors.
Consequently, HexaPro spikes may also ameliorate DNA or mRNA-based vaccines by producing more antigens per nucleic acid molecule, improving, in turn, the efficacy at the same dose, or by preserving efficacy at lower doses.
Finally, the authors highlighted a clear path towards the industrial level of production in order to meet global demand for this pivotal SARS-CoV-2 protein. Their work will undoubtedly facilitate the production of prefusion spikes for diagnostic testing kits and subunit vaccines, with potentially broad implications for next-generation vaccine design.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hsieh, C.L. et al. (2020). Structure-based Design of Prefusion-stabilized SARS-CoV-2 Spikes. bioRxiv. https://doi.org/10.1101/2020.05.30.125484.
- Peer reviewed and published scientific report.
Hsieh, Ching-Lin, Jory A. Goldsmith, Jeffrey M. Schaub, Andrea M. DiVenere, Hung-Che Kuo, Kamyab Javanmardi, Kevin C. Le, et al. 2020. “Structure-Based Design of Prefusion-Stabilized SARS-CoV-2 Spikes.” Science, July, eabd0826. https://doi.org/10.1126/science.abd0826. https://www.science.org/doi/10.1126/science.abd0826.