The novel coronavirus now termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes coronavirus disease 2019 (COVID-19) first appeared in Wuhan, Hubei Province, China, but has since spread over most of the inhabited world. SARS-CoV-2 is a single-stranded RNA virus, the seventh coronavirus known to infect humans. Among these, only the SARS, MERS, and SARS-CoV-2 are known to cause severe clinical disease in humans.
Variations in the Severity of COVID-19
Currently, the outbreak is known to have caused over 6.39 million cases and taken more than 383,000 lives the world over. However, huge gaps remain in the scientific understanding of how the virus causes a spectrum of disease ranging from asymptomatic to lethal respiratory failure.
At present, the incubation period of the virus is thought to be around 5 days, and almost 98% of cases become symptomatic within 11-12 days of infection. The precise percentage of asymptomatic infection is unknown, with estimates ranging from 5% to 80%. This has created several barriers to the containment of the disease.
The challenge to researchers today is to understand the pathogenesis and mode of transmission of the agent. One significant component of this is to identify the host and agent factors that are associated with COVID-19 severity. Among the known host factors are patient age, lymphopenia (reduced numbers of T cells), and hyperactivation of the inflammatory cascade, prominently IL-6, and IL-8.
Agent factors connected to the variation in the disease manifestations are less well recognized. At present, there are over 30,000 genomes available on various public databases like the Global Initiative on Sharing All Influenza Data (GISAID), many of which had accompanying patient data.
Using GWAS to Identify Genetic Variations

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Using this data, the researchers looked at 152 complete viral genomes from this database with the related patient data to identify the genetic variations that are potentially linked to the severity of COVID-19, via a genome-wide association study (GWAS).
The patient data associated with these genomes was clear enough to allow the patient to be classified as either symptomatic or asymptomatic. The investigators constructed the phylogenetic tree based on the maximum likelihood to visualize how these isolates were related to each other and to other sequences.
The study found that the isolates were very diverse, comprising 16 distinct lineages. However, symptomatic isolates came from almost every lineage, while most of the asymptomatic strains were from lineage B and B.5.
Again, asymptomatic viruses were from patients in Japan and India, while symptomatic isolates came from almost anywhere. In other words, 60/72 asymptomatic strains were from Japan, and 6 from India. On the other hand, 17/80, and 4/80 symptomatic strains were from Japan and India, respectively. The remaining 59 symptomatic strains came from a broad range of countries.
Variations at position 11,083 linked to symptomatic infection
The GWAS performed by the researchers was meant to pick up gene variants that might be linked to COVID-19 severity. They found genetic variations at position 11,083 to be significantly correlated with disease severity. This genomic position was repeated in 75% of all bootstrap trees.
Two variations were found at this site, thymine in about 49% and guanine in 47%, while in 5 sequences, the nucleotides were undetermined. The thymine variation was more likely to be found in asymptomatic infection and the guanine variant in symptomatic.
When the relative risk ratio was calculated for symptomatic disease, the G variant is 4.5 times more likely to be associated with symptoms than the T variant. The odds of symptomatic disease are 37 times higher for the former variant.
A small Shanghai study of 112 patients showed the T variant to be about twice as prevalent in asymptomatic patients compared to symptomatic cases, but not significantly. The current researchers attribute this to the small sample, and the clustering of mild and asymptomatic cases with severe cases, leading to a masking effect.
A second study identified this site as being subject to positive selection pressure, which agrees with the current study. The nearest genetic code to this genome is in the bat CoV, which has a G at this locus. This suggests that G is the original base at this site, prompting the current researchers to call this the 11083G>T.
This mutation is found in the non-structural protein nsp6, and causes the amino acid to change from leucine to phenylalanine.
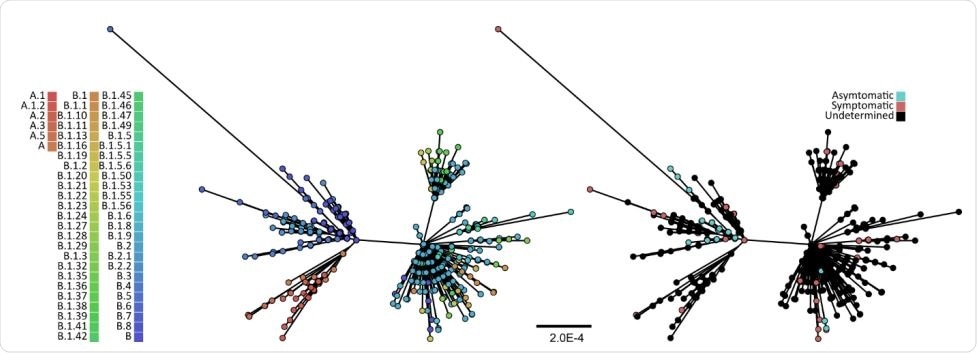
Maximum likelihood phylogeny of 625 SARS-CoV-2 full-length genomes. The tree was reconstructed using IQ93 TREE and the GRT+I nucleotide substitution model, the best-fit model as determined under the Bayesian information criterion by ModelFinder. The bootstrap clade support values were computed based on 1,000 pseudoreplicate datasets, and only branches with >70% bootstrap support are shown. The scales bar is in the units of substitutions per site. The tips were coloured either by their lineages (left) as identified by pangolin (github.com/hCoV-2019/pangolin), or by COVID-19 severity (right).
Variants cause variation in nucleotide-miRNA interaction
The interaction between the viral RNA and host microRNAs is thought to underlie the development of disease in viral infection. The present study examines the possibility that the T and G variants act differently because they bind to human miRNAs differently. The two miRNAs that are predicted to bind the G variant uniquely on the positive strand are miR-485-3p and miR-539-3p, both of which have the same nucleotide sequences at the 11,083 location.
The Biological Importance of The Mutation
Another study shows that miR-485 can reduce antiviral immunity through its interaction with the mRNA that transcribes the retinoic acid-inducible gene I (RIG-1). This gene gives rise to a protein that senses and responds to the presence of viral RNA in the host cell and triggers the antiviral response of the cell. The different ways in which these two variants bind to the miR-485-3p could result in different types of immune response and, therefore, different levels of severity of COVID-19.
The RIG-1 is a pathway that causes the production of the inflammatory cytokine TNF-α, setting off the uncontrolled inflammatory scenario that is called the cytokine storm. Therefore, another possibility is that the G variant might produce RNAs that preoccupy the miR-485-3p, as a result of which the RIG-1 pathway is expressed at an extremely high level and in an unregulated manner. This leads to massive overproduction of TNF- α and severe or critical disease. This theory needs more research to build up a picture of the interaction between these two elements.
The sequestration of miR-539-3p could also happen due to its interaction with the G variant, causing the increased expression of the Jagged1 factor that promotes new blood vessel formation. This miRNA can also increase the level of autophagy. It is interesting that the mutation of interest occurs in the nsp6 that is known to block host autophagy. This could hinder the ability of the cell to transport viral components to the lysosome to be broken down, resulting in a setting favorable for viral infection and making for a severe form of the disease.
What future developments can be expected?
The researchers comment: “Our results have potential applications for the development of better, and more informative test kits, potentially allowing for asymptomatic cases to be distinguished from symptomatic cases.” The use of bioinformatics has allowed hypotheses to be generated about the differential virus-host miRNA interaction in the two identified variants. The targeting of the G variant by human miRNAs could explain the variation in severity with these two variants. More research is required to understand how these variations are important in real life.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources