The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the ongoing pandemic of COVID-19, which has affected over 6.59 million people worldwide and caused almost 400,000 deaths in just over five months. Asymptomatic infection is thought to be extremely common with this virus, making containment measures extremely difficult.
Why serologic testing?
The identification of infected individuals before they become symptomatic, if indeed they do, is therefore of the highest importance to public health measures. Another huge benefit is the ability to identify those who are immune, which could facilitate selective lockdown application in many regions while still allowing the resumption of economic activity, without unduly endangering public health.
Thirdly, the identification of infected individuals in an accurate manner can help to understand the actual disease potential, risk factors, and transmission characteristics of the virus. Finally, serologic testing will be useful to determine how strong and how lasting natural immunity is. For all such testing, it is essential to develop rapid, inexpensive, and conveniently applicable methods that can be used at the site of care. At present, there are few such available.
Current assays and their limitations
One currently used point-of-care testing system is the lateral flow assay, which offers fast and easy testing on-site but is not sensitive or specific enough, besides lacking the ability to measure antibody concentrations. These are therefore inferior to laboratory-based test formats that use enzyme-linked immunosorbent assay (ELISA).
Currently, the weak link in the serologic assays now available is the test antigen used. The strongest antigen is the spike (S) glycoprotein, which is targeted by neutralizing antibodies and is essential for virus entry into the host cell. The nucleocapsid (N) antigen is also immunogenic but cross-reacts with antibodies against other coronaviruses.
The S1 subunit of the S protein is typically recommended for specific detection of an immune response to SARS-CoV-2. Suitable tests, according to the above criteria, using this antigen, are not presently available.
The MInD Procedure
The use of Magnetic Immuno-Detection (MInD) could overcome this issue. This is a procedure based on immunofiltration, where an antigen-coated matrix is used to trap specific antibodies in the sample applied to the matrix by gravity flow. These antibodies are then detected using secondary antibodies.
Finally, specially designed magnetic nanoparticles (MNPs) are added to label the secondary antibodies, the unbound antibodies are washed out, and the magnetic read-out is then obtained with a portable device that uses frequency magnetic mixing detection technology (FMMD).
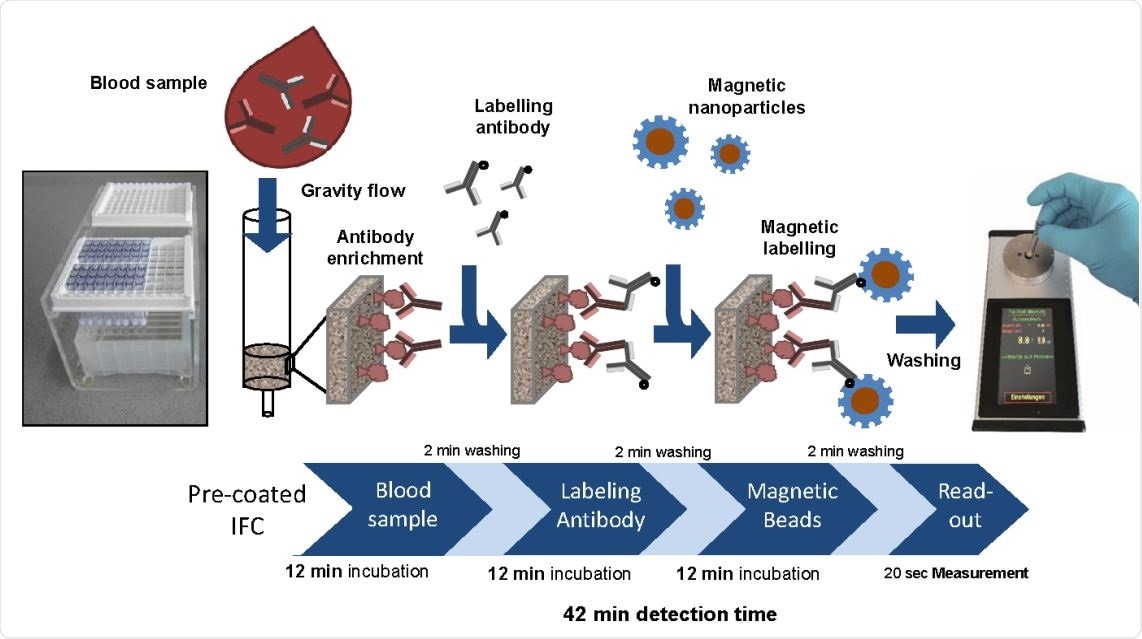
Proof-of-concept MInD assay setup using IFC coated with SARS-CoV-2 antigen. Assay steps and assay time are indicated. IFCs were coated with commercial SARS-CoV-2 S-protein peptide and blocked with BSA. Corresponding antibody was diluted either in PBS or spiked in human serum and applied to IFCs. Biotinylated secondary antibodies were added, followed by the application of streptavidin-functionalized MNP. Finally, IFCs were inserted into the portable magnetic read-out device. Measuring signal can be correlated to the amount of antibody in the sample and antibody titer can be determined. Assay time of this preliminary MInD setup was 42 min which is approximately four times faster than ELISA (161 min).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
How the Study was Performed
The current paper reports a proof-of-concept study of the use of MInD vs. standard ELISA, in which the former performs significantly better at specifically detecting SARS-CoV-2 antibodies in human serum.
The researchers used a peptide derived from the S protein of SARS-CoV-2 to coat the immunofiltration columns. Next, reactive antibodies to this peptide were introduced at varying concentrations into the columns and flushed through them. The specific antibodies bound to the coated antigen and were thus enriched in the column matrix.
They were then labeled using an isotype-specific biotinylated antibody. Then superparamagnetic streptavidin-functionalized magnetic particles were applied and flushed through the columns to label the secondary antibodies. After incubation, the excess MNPs were washed out, and the bound MNPs were detected using the FMMD-based portable magnetic reader.
The measuring signals correspond to the amount of bound antibody in the assayed sample.
Comparing the new method with laboratory-based ELISA, which was performed on the same sample as a reference point., threw up several observations.
Time-saving
The whole experiment took 161 minutes and can detect specific anti-SARS-CoV-2 antibodies at a range of 3.4 to 477 ng/mL. This is extremely sensitive with respect to the typical IgG levels in human serum, at about 10 mg/mL, and increasing when the individual is exposed to the antigen.
Higher range, broader sensitivity
The commercially available test kits for specific anti-S1-subunit antibodies can pick up IgG antibodies in three-fourths of samples within 10-20 days of infection. More sensitive assays might detect antibodies early in the course of infection.
However, when compared to the ELISA assay, the MInD assay was capable of detecting anti-SARS-CoV-2 antibodies in a range of 2.95 to 2040 ng/mL, which is five times broader. This means the test is far more sensitive than ELISA, with a lower detection limit, and a wider range of detection. It also allows quantitative measurements due to the broad dynamic range.
The procedure took only 42 minutes, which is only a quarter of the time required for the ELISA assay, which shows its potential for rapid testing. This can be further reduced by optimizing selected steps, such as reducing incubation time to 5 minutes or even less and using MNPs to which antigen-specific antibodies are already bound. This would eliminate the need for incubation with secondary antibodies.
With these adaptations, the total assay time would be below 20 minutes, which is comparable to that of a lateral flow assay.
Future Improvements and Implications
Other important innovations would be the use of antigens derived from the S1 subunit of the S protein, or multiple SARS-CoV-2 protein antigens in a mixture. Control antigens from other human coronaviruses should be tested to confirm the specificity of the enriched antibodies.
Multiplex testing using a range of MNPs could facilitate the detection of multiple subclasses of antibodies in a single assay. This could be useful in that it not only demonstrates seroconversion but also the phase of the infection, and the course of antibody formation.
Finally, optimizing the magnetic reader to a medical-diagnostics instrument will make the test suitable for such applications – such as including a barcode scanner and labeling the columns with patient-specific barcodes. The cost of the procedure is about a tenth or less of a typical ELISA, making it ideal for use in doctors’ offices.
The potential for the use of a single pipette for the assay procedure and the (perhaps battery-powered) magnetic reader device makes the device appropriate for various medical applications. For instance, it could be used in nursing and elderly care homes, or at airports, for fast and convenient point-of-care testing.
Once a vaccine is available, the same method can be used to monitor the emergence of antibodies and how long they last. Thus, this technique can be used in multiple ways and at different time points, as a fast and convenient point-of-care measurement without extra cost or sophisticated equipment.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Pietschmann, J. et al. (2020). Brief Communication: Magnetic Immuno-Detection of SARS-CoV-2 specific 2 Antibodies. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.02.131102. https://www.biorxiv.org/content/10.1101/2020.06.02.131102v1
- Peer reviewed and published scientific report.
Pietschmann, Jan, Nadja Voepel, Leonie Voß, Stefan Rasche, Max Schubert, Michael Kleines, Hans-Joachim Krause, Tamlyn M. Shaw, Holger Spiegel, and Florian Schroeper. 2021. “Development of Fast and Portable Frequency Magnetic Mixing-Based Serological SARS-CoV-2-Specific Antibody Detection Assay.” Frontiers in Microbiology 12 (May): 643275. https://doi.org/10.3389/fmicb.2021.643275. https://www.frontiersin.org/articles/10.3389/fmicb.2021.643275/full.