.jpg)
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of an apoptotic cell (blue) infected with SARS-COV-2 virus particles (red), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
A global spread of coronavirus disease (COVID-19) pandemic caused by SARS-CoV-2 made clear that we need to fully appreciate basic facts about the virus to make informed health care and policy decisions.
Initial data suggest that viral mutation rates are low; nonetheless, its large genome could facilitate insertions, deletions, and recombination events known to occur in other coronaviruses already in wide circulation.
Receptors and S-protein subunits
The entry of the SARS-CoV-2 genome into the cell starts when its surface subunit S1 of the spike glycoprotein (S-protein) binds to the ACE2 human receptor. At the same time, the S2 subunit is in charge of fixing the S-protein to the viral membrane surface.
Upon binding to the receptor, the S-protein is then primed by specific serine-protease (TMPRSS2), which leads to S-protein cleavage at S1/S2. After cleavage, S1 remains attached to ACE2, whereas subunit S2 anchors viral and cellular membranes – inducing fusion and viral entry.
But certain mutations in the viral genome can result in the complete absence of the S2 subunit responsible for such anchoring to the lipid membrane of the viral particle. The lack of the S2 anchor peptide indicates that S1 could be produced as a "free" protein (also known as free S1).
One alleged action of the secreted free S1 protein might be to bind the human ACE2 cell receptor, competing with whole viral particles to re-infect or newly infect respiratory tract cells – and resulting in less severe illness.
Basically, this could be explained as an effect of natural selection to weaken the infection and enable its persistence with minimal damage to the host, increasing the human-to-human transmission into the community.
Deep-sequencing of SARS-CoV-2 genes
This prompted the researchers from the Vall d'Hebron Hospital Universitari, Roche Diagnostics SL, and Universitat Autònoma de Barcelona in Barcelona, as well as from Instituto de Salud Carlos III in Madrid, to appraise naturally occurring SARS-CoV-2 gene deletions.
Upper respiratory tract specimens were collected for SARS-CoV-2 testing from individuals consulting in the emergency room of the Microbiology Department at the Vall d'Hebron Hospital Universitari in Barcelona, Spain.
The methodological approach involved deep-sequencing of the complete SARS-CoV-2 spike gene by utilizing 13 overlapping amplicons in laboratory-confirmed samples from 18 patients, of which 10 had a mild clinical presentation and 8 severe symptoms that necessitated ICU management.
The sequence analysis aimed to obtain high-quality haplotypes (i.e., a set of genetic determinants in an organism located on a single chromosome) fully covering the amplicons (a piece of DNA/RNA that is the source of amplification events).
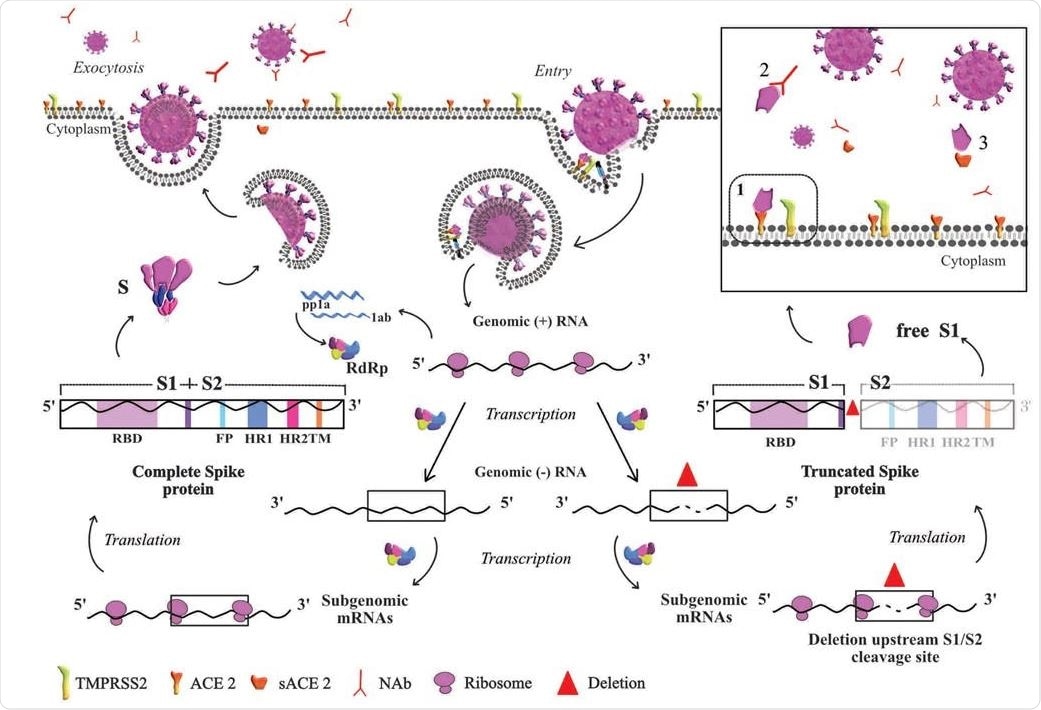
Based on the life cycle of SARS-CoV, this diagram represents the hypothesis derived from our results. Entry of the virus in the host cell is shown at the top right of the diagram. At the transcription step, two scenarios are depicted: to the left, the viral particle resulting from normal S protein, and to the right the viral particle resulting from truncated S. In normal conditions, once the nucleoprotein is freed into the cytoplasm ss+RNA is translated into the nonstructural proteins required for transcription. ss+RNA is transcribed into ss-RNA and later into genomic ss+RNA which is encapsidated (left side of the figure). Once the complete viral particle has been formed, it is secreted from the cell by exocytosis. The right side of the figure depicts the situation when a deletion occurs in the S gene during transcription of the complete genome and before subgenomic mRNAs are generated to produce the structural proteins. Translation of a deleted subgenomic spike mRNA would lead to a truncated S protein composed of the S1 domain without S2, which could be shed outside the cell as free S1. The box depicts possible destinations of free S1, which could bind to 1) the ACE2 cell receptor, 2) S1-specific neutralizing antibodies, or 3) free ACE2 receptor. The red triangle indicates the deletion in genomic RNA.
"Don't burn down the house"
This study described naturally occurring deletions in the SARS-CoV-2 S gene in patients with COVID-19. The deletions mainly clustered in two hot-spot regions; one upstream but rather close to the S1/S2 cleavage site, and the second situated upstream to the secondary cleavage site S2'.
The deletions were appreciably more prevalent in patients with mild than in those with severe forms of the disease – supporting the hypothesis that they may well be a strategy of natural selection used to decrease the injury caused after infection onset.
This strategic approach among viruses is also known as "don't burn down the house." More specifically, the propensity of the virus to bind with the ACE2 receptor and spread to others is unchanged, which means the transmission is enhanced by a mildly affected host.
Furthermore, the results suggest that the virus may generate free S1 protein that is subsequently released to the circulation, which may then compete with viral particles for the ACE2 receptor.
"Their effect on the protein could constitute a favorable regulatory mechanism emerging in the viral quasi-species to modulate the pathological effect of the infection," explain study authors.
Antiviral targeting
"To conclude, in-depth sequencing of the SARS-CoV-2 S gene in 18 patients with COVID-19 enabled the identification of a naturally occurring deletion very close to the S1/S2 cleavage site", study authors summarize their main findings.
Their results show that the mutant S would exert a significant impact on the S-protein, and also suggest that the virus could produce free S1, with further implications regarding the potential of S-protein as a target for vaccines and antiviral treatment approaches.
This groundbreaking insight into the biology of SARS-CoV-2 is definitely a big leap forward. In the meantime, it is pivotal to analyze further whether the free S1 is present in respiratory tract specimens and in plasma to guide further fundamental and practical research.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources