The use of antibodies produced in experimental animals or recovered from convalescent human plasma is an old but proven method of inducing immediate artificial immunity in severely ill patients suffering from infectious disease. The first such therapy was employed by Emil von Behring and Shibasaburo Kitasato against diphtheria.
The effectiveness of such plasma is dependent solely on the virus. For instance, convalescent human plasma failed to confer a survival advantage in the patient group as compared to the control group. Still, it was successfully used in the H1N1 pandemic of 2009 and the SARS outbreak. It has also been initiated in the current pandemic, and the results are hopeful.
Neutralizing Antibodies
Polyclonal antibodies derived from the serum of immunized animals or humans can neutralize the virus and thus aid the recovery of the patient. Other mechanisms are also possible, however, such as binding the Fc-gamma receptor and thus inducing phagocytosis, activation of the complement system, and antibody-dependent cellular cytotoxicity (ADCC).
The problem with using a convalescent serum is that it is impossible to standardize the composition or the effectiveness as it comes from different donors and is processed in different batches. Secondly, the risk of viral contamination is always alive and must be avoided. Thirdly, the neutralization potency must be preserved.
On average, the convalescent patient provides 400-800 mL plasma, and one treatment requires 250-300 mL plasma. Since each treatment needs two rounds, this limits the availability of donor plasma, at one donor for 2 patients at most.
A highly promising alternative is the development of human or humanized monoclonal antibodies, such as the molecule Palivizumab approved in 2009 for respiratory syncytial virus (RSV) infections.
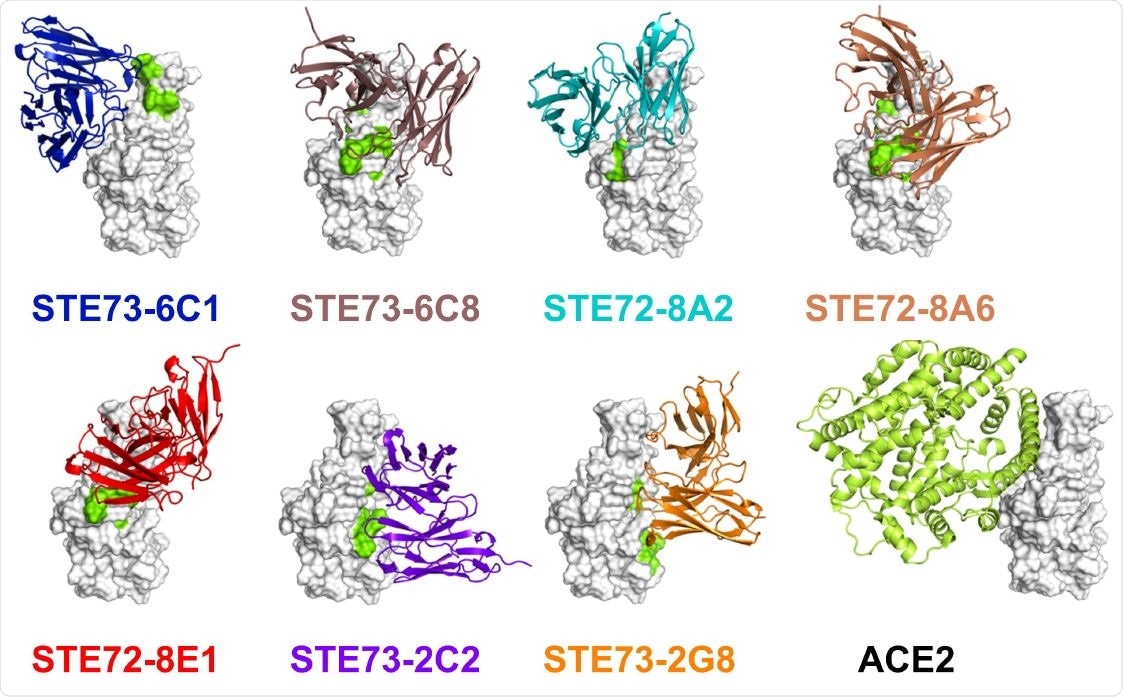
Epitope and structure modeling. A) Overview of the identified minimal epitope regions (MERs) for eight antibodies on RBD. Sequence SARS-CoV-2 (Gene bank QHD43416), SARS-CoV (Uniprot P59594). ACE2 receptor binding residues 74 are marked in yellow. B) Five of the inhibiting antibodies occupy the ACE2 binding region on the RBD (docking models based on epitope data). Two of them (right-most ones) bind to the opposite face of the RBD, suggesting allosteric inhibition. Experimentally validated computational models of the variable regions of the antibodies (colored cartoons) binding to the RBD (white surface, same orientation in all images) are shown. Amino acid residues recognized by each antibody in the peptide scanning experiment are marked in green on the RBD surface. The cartoon representation of ACE2 is also shown for comparison.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Human Monoclonal Antibodies
The current study aimed at producing human monoclonal antibodies that can neutralize SARS-CoV-2 from a universal gene library generated from healthy donors before the pandemic. The advantage of this approach is that human antibodies against this virus can be chosen against COVID-19 individuals. Phage display derived antibodies are already an established class of medications, with 12 already approved.
The antibodies capable of blocking the interaction of the viral spike protein with the human ACE2 receptor were first selected, as only these can neutralize the virus if it follows the pattern of the earlier SARS-CoV. Using the S1 subunit protein generated in insect cells, which was better at antibody selection than the mammalian equivalent, the researchers found 309 unique and completely human antibodies.
From these, over a hundred were recloned and tested for their ability to inhibit the binding of the S protein trimer with ACE2 on host cells, using flow cytometry. Some of them were able to inhibit the binding at a 1:1 ratio or even better, for the antigen-binding site: spike monomer/RBD. Since the RBD has to be in the less stable “up” conformation in order to bind to the ACE2, and since different RBDs on the same trimer may be in different conformations, this could explain the below 1:1 effective molar ratio of antibody to spike.
The antibodies are better at preventing RBD than S-trimer binding to the ACE2 receptor, possibly because of their higher affinity for the former, caused by the presence of entirely or partly inaccessible epitopes on the spike protein.
The strongest inhibition of binding was seen on human lung cells Calu-3, which are more similar to the living cells than experimental cells that transiently express ACE2 at high levels. The latter was first used to evaluate inhibiting potency more accurately.
Combinations of antibodies are synergistic and may also avert escape mutants. Here too, the best combinations were markedly better when antibody excess was used at a 30:1 molar ratio of antibody to antigen.
All of the 17 antibodies were tested in neutralization assays with SARS-CoV-2, producing an apparent measurable effect at a low titer. This allowed the researchers to select the best antiviral antibodies rapidly. Some antibodies did, however, increase the binding of the virus spike protein to the ACE2-bearing cells and lead to increased infectivity – possibly by multimeric spike formation or by stabilizing a conformation that promotes infection.
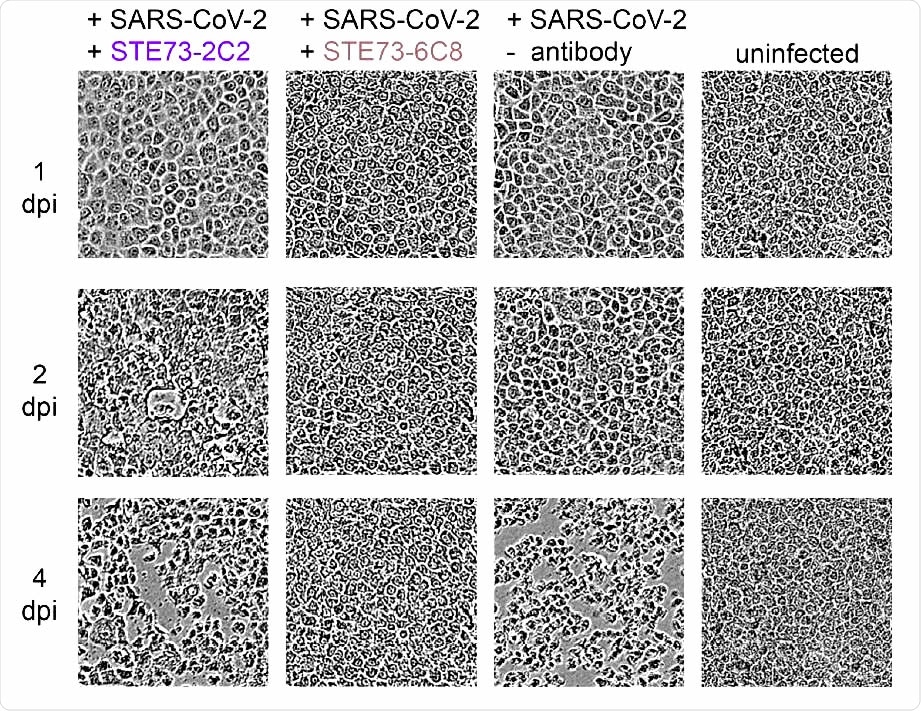
SARS-CoV-2 neutralization. Neutralization analysis using 250 pfu of SARS-CoV-2 in a CPE based neutralization assay. A) Cell monolayer occupancy at 4 days post-infection in the absence of neutralizing antibodies was compared to uninfected control cells and median values were normalized as 0 and 100% occupancy, respectively. Histograms indicate medians of normalized monolayer occupancy in a neutralization assay using 1 µg/mL (~10 nM) antibody for each of the 17 tested antibodies. Black dots indicate monolayer occupancy in individual assays (4-6 measurements per sample). B) Representative phase contrast microscopy pictures of uninfected cells, cells infected in absence of antibodies, in the presence of a poorly neutralizing antibody (STE73-2C2) or of a highly neutralizing antibody (STE73-6C8).
Implications and Applications
Such antibodies can be developed for passive immunotherapy, to prevent infection in groups like healthcare workers who are at high risk, and to modulate the severity of infection in sick individuals. The risk of antibody-dependent enhancement (ADE), which could worsen the severity of the disease, has to be weighed carefully before using them clinically. This could be minimized by using only RBD or silenced Fc parts, without the capability for Fc-gamma or C1q binding.
The researchers point out the advantage of this approach to generating neutralizing antibodies: “Universal libraries from healthy donors offer the advantage that antibodies can be generated quickly and independent from the availability of material from recovered patients in a pandemic situation.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources