As the COVID-19 pandemic caused by the coronavirus severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) continues to spread around the world, many researchers are studying epidemiological models to predict its propagation. As of yet, there are no effective vaccines that could counter the SARS- CoV-2 infection or drugs to treat COVID-19 disease.
Now, a new study shows that one of the enzymes of the virus – a protease called Mpro, could be a target that could be utilized to kill the virus. The study titled, “Topological analysis of SARS-CoV-2 main protease,” was released prior to peer-review on the online site bioRxiv and is to be published this week in the journal Chaos.
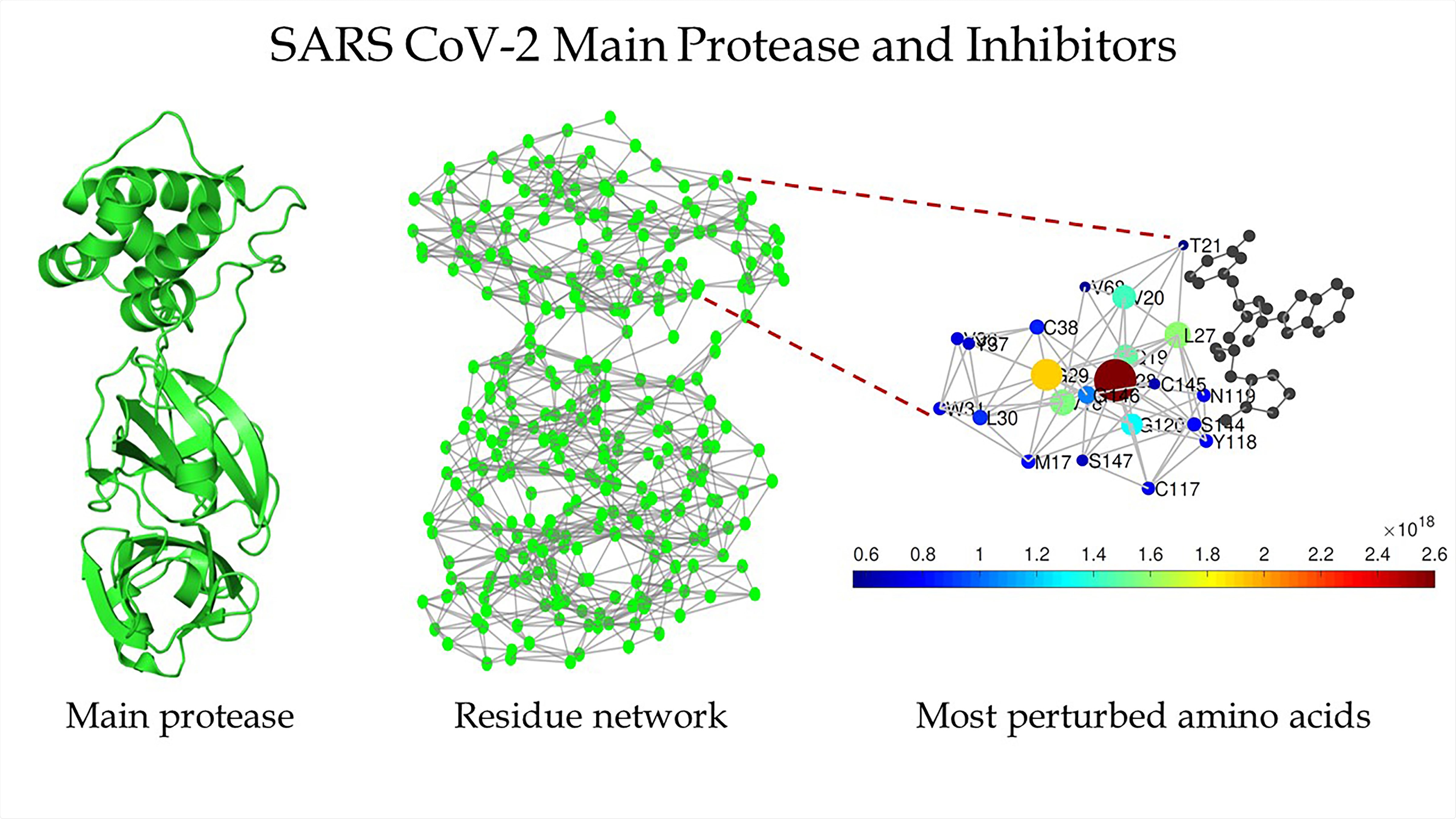
Schematic of the main protease of SARS-CoV-2 (left), the protein residue network of the main protease of SARS-CoV-2 (center), and a zoomed-in view of the region around the binding site as detected by Estrada (right). CREDIT Ernesto Estrada
What was this study about?
The researchers wrote that the SARS-CoV-2 belongs to the genus Betacoronavirus. They explained that the SARS (SARS CoV-1) and the MERS virus before this had caused prior respiratory disease epidemics and that the novel SARS-CoV-2 shares about 82 percent of its genome with SARS-CoV-1. They explained that this was the reason that the earlier antiviral drugs such as Remdesivir or the antimalarials Chloroquine and Hydroxychloroquine, were being repurposed and tested as COVID-19 treatments. They wrote that specific therapy against the virus is still elusive.
The team of researchers wrote that earlier drugs had targeted viral proteases in the case of the human immunodeficiency virus (HIV) and SARS-CoV-1. They call these proteases an “attractive pharmacological target.” SARS-CoV-2’s main protease (Mpro) which is the main enzyme that allows it to process the polyproteins by proteolysis - breaking up of the proteins. They speculated that if the activity of the Mpro could be blocked, viral replication could also be stopped.
What is the role of the protease?
The team led by Ernesto Estrada, a mathematician, an expert in complex systems of the ARAID Foundation at the University of Zaragoza, suggests that the protease is specific for the virus and is absent in humans. This would mean that if targeted, this drug would leave the host cells unaffected and reduced the risks of toxicities. The team wrote that in an earlier study, another group of researchers led by Zhang had already found the three-dimensional structure of the CoV-2 Mpro. The size of the protease is 1.75Å (Angstrom). They wrote that the structure of the protease CoV-2 Mpro was similar to another molecule - α-ketoamide inhibitor. This has also been elucidated by other researchers, they wrote.
This protease they wrote has an amino acid sequence that has 96 percent common with the protease of CoV-1. Of the 303 amino acids, they explained, only 12 were different. The difference between the two was only 0.53 Å. “It seems like the two proteases are almost identical in their three-dimensional structures.”
Estrada said, “I noticed that chemists had already found some potent inhibitors of the main protease of SARS-CoV-2 and that they'd resolved the structure of this protein via X-ray crystallography. It was shocking to see that this protease is very similar to that of the SARS coronavirus, which produced the epidemics of 2003, SARS-CoV-1.” He added, “If you line up the amino acid sequences of both proteases, there are only 12 out of 306 residues that don't coincide. Is there something hidden behind these apparent similarities between the two proteases? Can we learn something from them to improve the design of drugs against the virus?”
“They're called protein residue networks, where we represent every amino acid as a node, and the interaction between two amino acids is represented by a link between the two,” he explained.
The hypotheses raised included if the similarities between the two proteases could also mean “topological similarities.” They wrote that topological similarity would mean “network theoretic representation of a protein.” They explained that in this network of the protein structure, there is a network that “represents amino acids and the edges connecting them indicate that the corresponding residues are at a distance in which they can interact with each other.” They wrote that this calculation could get the importance of the structural and dynamic of an amino acid within the protein. For this study, the team thus constructed protein residue networks (PRN) for CoV-2 Mpro as well as for its inhibitors.
What was found?
The results of the study show that both the proteases of CoV-1 and 2 hare similar in terms of topological characteristics. They found that the difference was not more than 4 percent. Transmissions, however, can alter the proteases they wrote. They wrote, “CoV-2 Mpro is 300% more sensitive than CoV-1 Mpro in transmitting tiny structural changes across the whole protein through long-range interactions.”
These alterations of the amino acids on the Mpro are found in the catalytic site Cyst-145, where the inhibitors can bind. They found that the most important amino acids in the protease structure were also the most important ones in the binding of the inhibitors with the CoV-2 Mpro. This, they wrote, could play a role in the drug design that could inhibit the protease.
Speaking on the study, Estrada said, “But a couple of years ago, we developed a more sophisticated mathematical measure that allows us to detect how far away a perturbation within a network can be propagated. That work was of a very theoretical, mathematical nature, but we had speculated that it could be useful for the study of proteins.” He added, “This means that when a protein is perturbed, for instance, by water within the intracellular environment, such perturbations are transmitted through a network of intraresidues that form the 3D structure of the protein. If such perturbation is produced around a given amino acid within the protease of SARS-CoV-1, it's transmitted only through a close environment around that perturbed amino acid.”
Conclusions and implications
Estrada said, “It's remarkable because it means that with tiny structural differences, the protease of SARS-CoV-2 is much more effective within intra-residue communications. It should be much more effective in doing its job as a proteolytic enzyme of the virus. The devil did a nearly perfect job here, but he left the door open. This great sensibility of the SARS-CoV-2 protease to perturbations can be its Achilles' heel in relation to inhibitors.”