The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the cause of the ongoing COVID-19 pandemic. As of June 10, 2020, it has caused over 7.13 million cases and more than 406,000 deaths. In India, the outbreak started slowly and late but has already caused over 267,000 cases and 7,700 deaths.
The mortality recorded so far indicates a case fatality rate of about 3.4%, which is lower by far than that of either the SARS or the MERS outbreaks. On the other hand, the current pandemic is due to a strain that spreads much faster than either of the earlier outbreaks.
In the absence of any effective vaccine or therapeutic drug, scientific teams all over the world are working hard to understand the viral genome, the pathogenesis of the disease, and factors that contribute to the severity of the disease. The occurrence of mutations is important since these could potentially disrupt the efficacy of any vaccine or tailored therapeutic agent by altering the antigenic structure of the virus.
The Gujarat Biotechnology Research Centre (GBRC), Gujarat, India, is among the many institutions involved in viral sequencing. It has already completely sequenced over 144 SARS-CoV-2 strains circulating in the state, using samples from the state testing center. The study shows certain unique mutations in Indian isolates.
Mutations in One Indian Strain
The current paper describes two samples from a married couple aged 66 years, who contracted the infection from their son, who in turn acquired it via community spread. The infection in all cases was mild. The genome sequence obtained from both husband and wife had a size of about 29,902 bp, which corresponds to a single nucleotide deletion compared to the Wuhan strain.
The single deletion occurred at different locations in these strains. Moreover, there were ten mutations overall in each genome with respect to the Wuhan strain.
Analysis of the mutations showed that single nucleotide mutations led to changes in amino acids, possibly causing a conformational change in the viral structural protein.
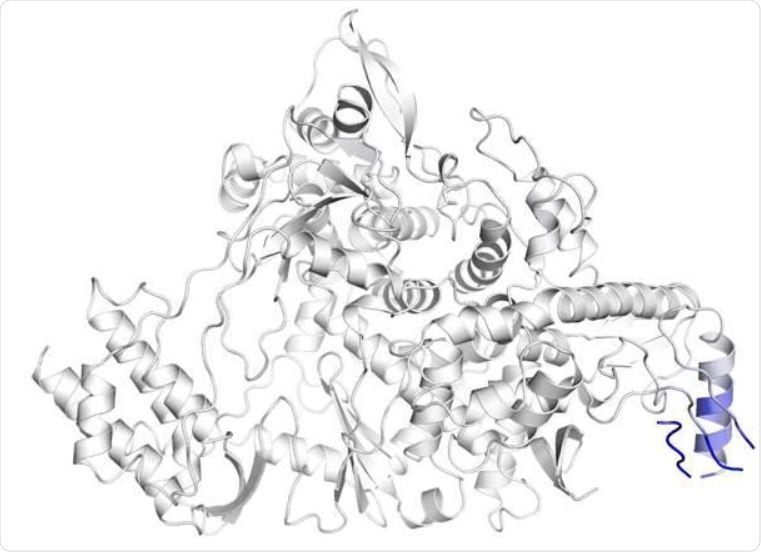
The substitution of proline with leucine at 323rd residue of RdRP substantially changes the inter-atomic interaction of the amino acid residues around the mutation site. The effect of the mutation on the structure of RdRP has been shown where blue represents a rigidity in the structure and red represents a gain in flexibility. (b and c) The inter-atomic interactions of the residues in the vicinity of 323 rd amino acid of wild-type RdRP protein and P323L mutated RdRP protein, respectively.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Mutations and Their Effects
In both patients, there was a point mutation of C241T in an untranslated region at the 5’ end, which has a negligible chance of producing significant effects on the replication of the virus.
Another mutation is in the protein ORF1ab, namely, C1059T, which changes the uncharged polar threonine into the hydrophobic isoleucine. Threonine interacts through hydrogen bonds with the polar amino acids deep within the protein structure. This may help form a bend in the protein because of the relative positions of glycine and proline.
Isoleucine could, however, disrupt such a bend because it cannot form hydrogen bonds, and could, moreover, promote hydrophobic interactions with phenylalanine residues around it. Nevertheless, in the absence of firm knowledge about the non-structural protein nsp2 or any of its homologous structures, no structural insights could be made.
The native nsp12 binds with the cofactors (including nsp8) in the RNA dependent RNA polymerase (RdRP) complex. The Indian strains show the C14408T mutation in this binding region, which substitutes proline with threonine. The proline causes an anomalous turn in the alpha-helix that houses it, which is also one of the nsp12-nsp8 interaction sites.
This changes the alpha-helix structure and may disrupt the binding of RdRP to nsp8, inhibiting its function. Thus, the mutation could stabilize this region, make it very rigid and thus inhibit the proper function of RdRP, which might require substantial flexibility.
Another nsp12 mutation, C15371T, substitutes the polar threonine with the hydrophobic methionine residue, which also leads to a change in the structure of nsp12. The paired cysteine residues near threonine promote cystine formation via cellular oxidation. Cysteine residues bound by disulfide bridges have significant enzymatic and catalytic activity, which plays a significant part in the folding and stabilization of the extracellular proteins in contact with a more hostile extracellular milieu.
This mutation, therefore, could disrupt the formation of such disulfide bridges and consequent enzymatic activity. The methionine side chain occupies significant space and disturbs the protein 3D structure and stability. Methionine will sequester within the protein due to its hydrophobicity, which reduces the reactivity of the neighboring cysteine residues as well. On the other hand, the location of the mutation, in a flexible loop region distant from the RdRP catalytic site, reduces the chances that the protein will undergo any significant structural change due to the mutation.
The same applies to the C17747T mutation, which substitutes proline into leucine in nsp13 (a helicase), at a distance from the functionally active site, and in a flexible loop region, reducing the chances of significant disruption of enzyme activity.
The A23403G is an aspartic acid to glycine substitution, causing an S protein mutation in the loop between two successive antiparallel beta-strands. The substitution by the flexible glycine could disrupt the strand structure of one of the four strands in the region. Since these strands take part in spike protein trimerization, required to bind the S protein to the ACE2 receptor, this might inhibit trimer formation, by reducing the binding affinity of these components.
Other mutations include G25563T (glutamine into histidine in the ORF3a protein), possibly converting it to a proteolytic cleavage site but perhaps leaving the corresponding viroporin function undisturbed.
Mutations that Affect Viral Replication and Virulence
The G28221T substitutes a stop codon for glutamic acid, so that the C-terminal of ORF8 is truncated by the deletion of a 36-nucleotide long segment. In essence, this corresponds to the cleaving off of the fourth beta-strand in this region, probably disrupting the function of the protein. Interestingly, earlier researchers reported the effect of a 29-nucleotide stretch in the SARS-CoV-1, which was the most apparent mutation in the early phase of its spread among humans.
Importantly, this 29-nucleotide deletion led to a 23-fold reduction in viral replication, indicating its attenuating effect on the virus. This may help explain why the Indian strains seem to be less virulent compared to others and may indicate that this strain should be studied for its potential in vaccine development.
Finally, the G28371T mutation substitutes serine into the hydrophobic residue isoleucine, in a region of the N protein distant from any functional domain and, therefore, unlikely to produce much effect on the structure of the protein. However, this is surrounded by a site for glycosylation, which is an essential post-translational modification of the protein that confers virulence. This mutation could, therefore, have a marked impact on viral protein glycosylation.
The study identifies some possible reasons for the loss of virulence in Indian strains and offers potential avenues for vaccine development.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.