Angiotensin-converting enzyme 2 (ACE2), expressed by cells in many human organs and tissues, has already been established as the functional host receptor for SARS-CoV-2 – the virus responsible for the ongoing devastating global pandemic of coronavirus disease 2019 (COVID-19).
SARS-CoV-2 spike glycoprotein (often abbreviated as S-protein) is a pivotal trimeric viral structure with either none or one of three receptor-binding domains (RBDs) in the "up" state, capable of binding to its target in order to infect the cells.
Practically all patients who recover from COVID-19 produce IgM and IgG antibodies against the S-protein; hence, a large number of serologic tests have been developed and marketed in order to appraise the immune response. Nonetheless, it is still unclear how effective the detected antibodies actually are at neutralizing the virus.
In any case, adequate antiviral immunity to SARS-CoV-2 will definitely be needed to return to normalcy from the current pandemic. Unfortunately, we still do not know whether the infection or vaccination will lead to adequate vigorous and long-lasting immunity.
Likewise, we urgently need high throughput serological tests that can be used to establish the presence and functional value of anti-SARS-COV2 antibodies to capture the full repertoire of inhibition of viral infectivity.
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of a cell (purple) infected with SARS-COV-2 virus particles (yellow), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Consequently, researchers from Seattle Children's Research Institute and the University of Washington in Seattle, US, described a novel, cell-free assay to quantify the ability of human plasma samples to impede the binding of ACE2 to the recombinant trimeric S-protein.
Microspheres and flow cytometry
In a nutshell, their methodological approach was characterized by the utilization of microsphere-based flow cytometry assays that successfully quantified anti-spike IgG antibodies in human plasma, as well as the ability to inhibit the binding of spike protein to ACE2.
Basically, the researchers optimized immunoprecipitation-flow cytometry assays to detect the appearance of anti-SARS-CoV-2 spike antibodies in human sera by covalently coupling either a spike trimer construct or the recombinant RBD fragment to carboxy-modified polystyrene latex or Luminex MagPlex microspheres.
Plasma specimens for this study were obtained from participants with PCR-positive nasopharyngeal secretions to COVID-19 more than fourteen days after symptom onset. Negative controls consisted of banked samples that were collected from healthy individuals prior to January 2020.
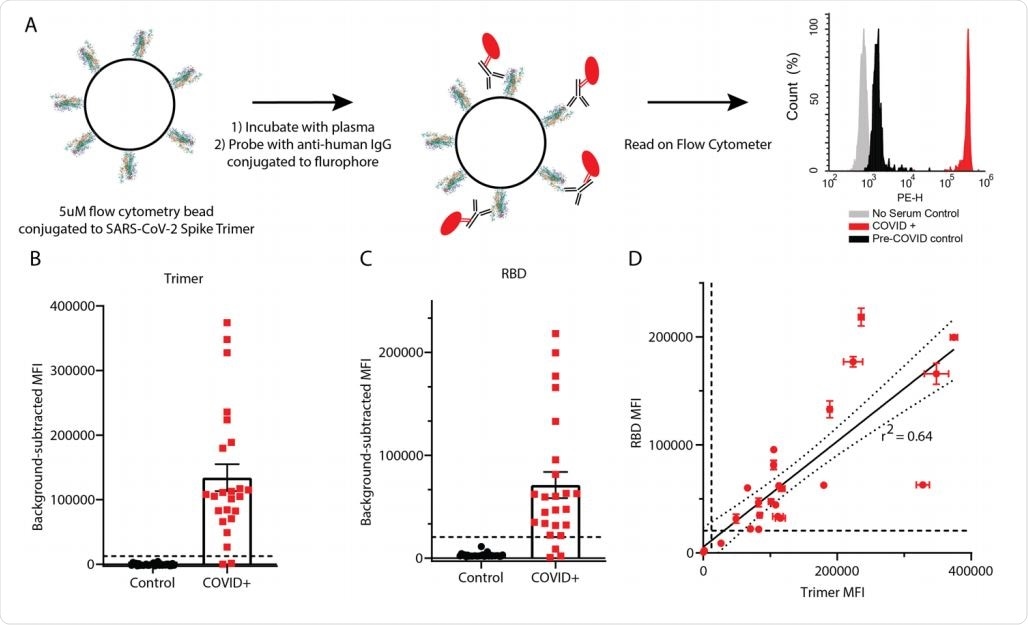
Detection of anti-SARS-CoV-2 IgG using RBD and trimer constructs. (A) Graphical representation of the methods. (B, C) IgG levels in plasma from 24 COVID+ and 30 pre-COVID-19 subjects were measured using trimer-conjugated (B) or RBD-conjugated (C) microspheres. Dashed lines indicate the cut-off for positive sample designation, calculated as (Maximum control value + 5 standard deviations, at MFI = 12,432 for trimer, 27119 for RBD. E) The median fluorescent intensity of IgG measured on the trimer and RBD assays was significantly correlated. Dashed lines indicate cut-offs for positive designation, and the solid line shows a linear regression with 95% confidence intervals indicated with dotted lines, r2 = 0.64, slope is significantly non-zero (F1,46=82.4, p<0.0001).
Trimer-reactive immunoglobulins as correlates of SARS-CoV-2 protection
This study brings the first description of a correlation between disease severity and the level of antibodies in a community cohort. Findings suggest that systemic illness or an inflammatory response may lead to higher antibody titer, although both the presence of fever and antibody titers may reflect intrinsic host differences.
More specifically, among the 24 study subjects with COVID-19, the presence of fever was linked to higher levels of anti-trimer IgG antibodies, as well as subsequent inhibition of binding to the human ACE2 receptor.
Importantly, a decrease in trimer-reactive immunoglobulins from plasma reduced ACE2-trimer inhibitory capacity much more than just the depletion of RBD-reactive immunoglobulins – suggesting that inhibitory antibodies act by binding both within and outside of the RBD.
"While we identified robust trimer-ACE2 inhibition in IgG-positive post-convalescent plasma, only weak inhibition of RDB-ACE2 binding was detectable, and only in the minority of samples", study authors further explain their findings delineated in medRxiv paper.
Additionally, no correlation was observed between anti-RBD levels and the inhibition of ACE2-RBD binding, hinting that certain individuals may mount an antibody response to the RBD that is unable to inhibit binding of the RBD construct to the recombinant, bead-bound ACE2.
Towards the routine clinical evaluation of functional immunity
"Our data, demonstrating in vitro neutralization of ACE2-trimer binding in 92% of recovered patients, adds to the growing literature that collectively suggests that infection with SARS-CoV-2 results in robust immunity, at least in the short-term", explains study authors.
Regardless of the underlying mechanism, this study shows that (at least for practical purposes) the trimer construct gives a reliable quantitative readout of immunity. In contrast, the RBD construct is less useful in that regard.
"The trimer inhibition assay presented here could be broadly useful in the settings of routine clinical evaluation of functional immunity in recovered patients, selecting the most potent post-convalescent plasma for use as therapy, and evaluating the functionality of antibodies produced in response to experimental vaccines currently under development," conclude study authors.
Future studies should use trimer-style constructs for serological tests, but also address other potential antigens or auxiliary immune mechanisms that might prevent viral entry into the cell, while older or more severely ill subjects should be included in the case-mix.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gniffke, E.P. et al. (2020). Plasma from recovered COVID19 subjects inhibits spike protein binding to ACE2 in a microsphere-based inhibition assay. medRxiv. https://doi.org/10.1101/2020.06.09.20127050.
- Peer reviewed and published scientific report.
Gniffke, Edward P, Whitney E Harrington, Nicholas Dambrauskas, Yonghou Jiang, Olesya Trakhimets, Vladimir Vigdorovich, Lisa Frenkel, D Noah Sather, and Stephen E P Smith. 2020. “Plasma from Recovered COVID-19 Patients Inhibits Spike Protein Binding to ACE2 in a Microsphere-Based Inhibition Assay.” The Journal of Infectious Diseases 222 (12): 1965–73. https://doi.org/10.1093/infdis/jiaa508. https://academic.oup.com/jid/article/222/12/1965/5892952.