The ongoing COVID-19 pandemic has caused well over 7.84 million cases and 431,000 deaths, worldwide, in less than six months. The disease is caused by a novel coronavirus now named the SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), which results in mostly asymptomatic or mild infection. However, in about 20% of patients, the virus can cause severe or even critical disease, including pneumonia, respiratory failure, and death.
Unfortunately, there is neither an effective vaccine nor have any therapeutic drugs been discovered so far. Thus, scientists are still searching for clues to the effective treatment and prevention of this infection.
Many infectious agents cause asymptomatic disease in a high percentage of individuals, and even in infected people, there is significant variability in the symptoms. This is true of COVID-19 as well. The search for predictors of severe disease has been going on, and some studies show that inflammatory mediators are higher and the counts of lymphocytes lower in those with severe disease.
The question is, what determines the host’s response to the infecting virus? Some researchers have reported that genetic factors play a significant role in this process, judging by the host response to other viruses like HIV, HBV, HCV, SARS-CoV, and the influenza virus. The HLA genes, interferon-stimulating genes (ISG), and other genes that affect viral replication all show that the genetic background partly determines, at least, how the organism manifests the infection.
Genetic Factors Predicting Severe COVID-19
A UK twin study shows that heritability characteristics for self-reported symptoms and the predicted onset of disease comprise 30% to 50% of the variability in the manifestation of and susceptibility to the disease. A recent large scale international study showed that blood group O was linked to a lower risk of COVID-19 and blood group A to a higher risk.
Other researchers have studied mutations in important genes like ACE2 and TMPRSS2, and genome-wide association studies (GWAS) indicate some other genes. Meanwhile, SARS-CoV-2 genomes are being sequenced, and the sequences shared worldwide through international databases.
The Study
Now, a new paper published on the preprint server medRxiv* reports the results of an analysis of the association between the genetic variants in the patients and the clinical progression of the disease. The study included 332 patients hospitalized with COVID-19 in a single-center, with widely varying laboratory and clinical features. There were 25, 12, and 17 patients with asymptomatic, mild, and critical disease. The largest number had moderate symptoms (225), followed by severe disease in 63 patients.
Asymptomatic, mild, and moderate disease refers to the presence of no or mild pneumonia. Severe and critical disease refers to impaired lung function and blood oxygenation, with multi-organ dysfunction, septic shock, and respiratory failure.in the latter group.
Risk factors for severe and critical disease include older age, longer duration of disease, male sex, and underlying disease conditions.
The researchers carried out deep whole-genome sequencing, which allows the effects of even rare and loss of function variants to be estimated. The process allowed for the detection of over 22 million variants, both common and rare.
Using these variants, the investigators then related them to the host factors using GWAS with both single variants and genes. They also examined the difference in frequency of the alleles that led to the truncation of proteins, and the HLA alleles, in the patient groups. They also called other publicly available and selected genomes to aid their search for potential genes that affect the genetic susceptibility to this infection.
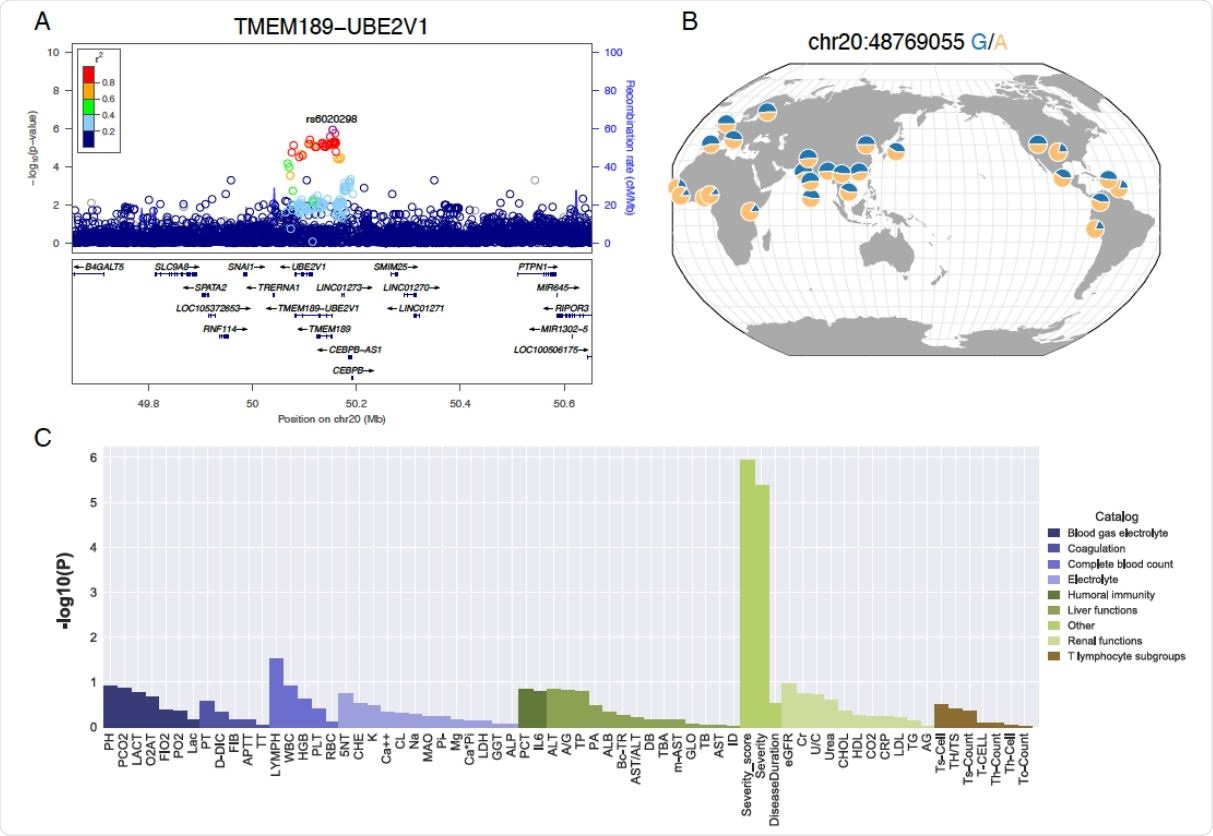
LD, allele frequency and pleiotropic effects of the TMEM189-UBE2V1 signal suggestively associated with COVID-19 patient severity. A) Locuszoom plot shows the p-value of the SNPs centering the lead SNP rs6020298 and the recombination rate. Color of the dots indicate linkage disequilibrium r2 metric. B) Allele frequency of s6020298 among the 1000 genomes populations. The allele frequency of the reference and alternative allele is visualized by the geography of genetic variants browser developed by the university of Chicago. C) P-value of the single variant genome-wide association test for the sixty-four laboratory assessments at the lead SNP rs6020298.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
No Single Gene Predicts Disease Manifestation
The most important finding from this study is that changes in a single gene do not predict the clinical severity of COVID-19. The researchers found almost 4,900 mutations predicted to cause loss of function, for an average of 200 such per patient. Severely ill patients typically had a more significant number of loss of function insertions compared to the mildly ill group.
When they narrowed down their search to mutations involving loss of function or missense variants for the ACE2 receptor, the TMPRSS2 protein primer gene, and other genes that are predicted to play a role in the interaction of host and virus, no significant association was found. A missense mutation in TMPRSS2 is actually less common in critically ill patients than in others. This may be because the substitution of valine by methionine causes instability of the TMPRSS2 and reduces viral binding to ACE2.
.jpg)
ACE2 acts as a cellular doorway – a receptor – for the virus that causes COVID-19.. Image Credit: Kateryna Kon / Shutterstock
Study of the HLA genes associated with severe disease showed that a higher prevalence of the haplotype HLA-A*11:01, 341 B*51:01, and C*14:02. This comprises three HLA alleles with strong linkage equilibrium and which were individually significantly linked to severe disease compared to mild manifestations.
A gene locus related to cytokines in the IL-1 pathway contains one risk allele that is more common in the severely ill patients and may be potentially related to the clinical manifestation of COVID-19.
Limitations and Implications
The study is limited by the lack of power to identify significant genome-wide genetic variants that have minor allele frequency of more than 0.2, because of the small size of the sample. The low percentage of asymptomatic patients is another factor that requires much more work to find the host factors more prevalent in individuals who have successfully resisted symptomatic COVID-19.
The researchers sum up: “Our results highlight several genetic factors involved in the immune responses. The summary statistics will encourage international collaborative efforts to understand the host-pathogen interaction and to contain the COVID-19 outbreak.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources