The current pandemic of COVID-19 has taken over 432,000 lives, with well over 7.89 million confirmed cases worldwide as on June 15, 2020. This new coronavirus is primarily a respiratory virus, causing mostly asymptomatic and mild disease. In a significant minority, the virus induces severe lung symptoms and multi-organ damage, which is often fatal.
Viral Entry Mechanism
The virus enters the host cells by binding to the human angiotensin-converting enzyme (hACE2) receptor on the cell membrane. This is mediated by the spike protein of SARS-CoV-2, a transmembrane protein, which mediates the recognition of the ACE2 receptor and membrane fusion, necessary for viral entry into the host cell.
The S-protein exists as a trimer on the viral membrane, each protomer consisting of two functional subunits, S1 and S2. These are responsible for viral-receptor binding and virus-host cell membrane fusion, respectively. The S1 subunit bears the receptor-binding domain (RBD) on its C-terminal domain.
X-ray crystallography shows that the RBD acts like a hinge or a flap, with an ‘up’ or ‘down’ conformation. In the latter, the amino acid residues that bind the ACE2 molecule are hidden; in the former, exposed.
The RBD itself contains the receptor-binding motif (RBM), which actually comes into contact with the carboxypeptidase domain of the ACE2 molecule. This is a variable region of the RBD compared to the rest of the protein, which raises the question of whether the other coronaviruses like SARS-CoV can create cross-reactive antibodies to SARS-CoV-2. The rest of the RBD makes the RBD more stable.
The entry of the virus into the host cell requires the spike or S protein to be cleaved in two steps. The first step occurs following the binding of the virus to the ACE2 protein and is accomplished by cellular proteases acting at the region between S1 and S2, namely, the transmembrane serine protease TMPRSS2.
There is also a unique furin cleavage site in this region, not found in SARS-CoV. This makes the virus more adaptable for cleavage by other cellular proteases as well, thus making it able to infect more cell types, since furin proteases are widely found, particularly in the respiratory tract.
This cleavage step is required for the initiation of membrane fusion by the S2 subunit, and this involves a second site of cleavage on this site, which is now considered essential to activate the S2 protein.
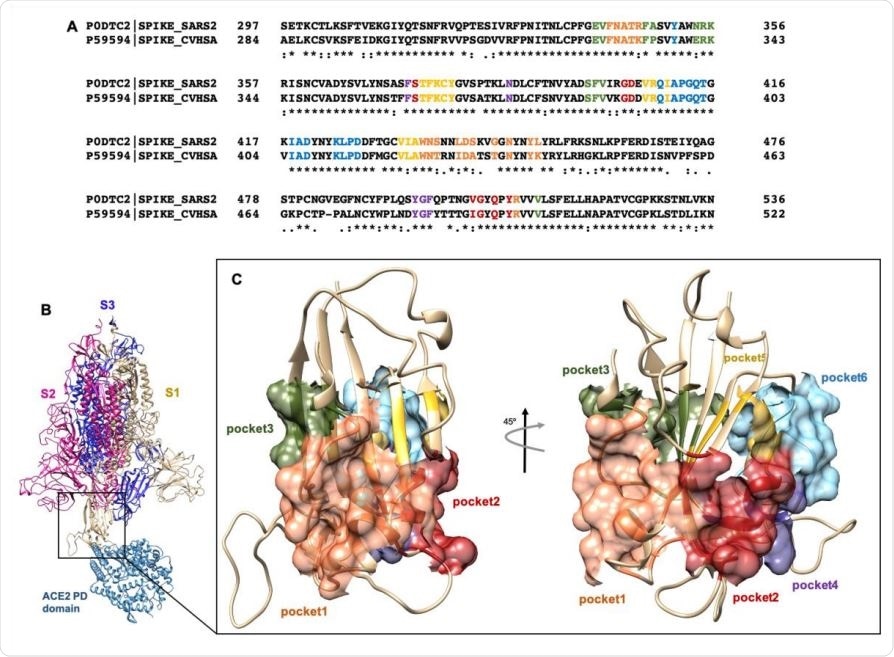
(A) Clustal Omega alignment of RBD regions of SARS-CoV and SARS-CoV-2 Spike protein. Residues bearing to different pockets are colored respectively yellow (Pocket 1), green (Pocket 2), light blue (Pocket 3), magenta (Pocket 4), red (pocket 5) and dark slate blue (Pocket 6). (B) Illustration representation of the trimer of SARS-2 Spike protein in complex with the PD domain of ACE2. Complex obtained through the superposition of the PDB structures 6VSB and 6M0J. (C) Surface representation of the six selected pockets used for the screening.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Drugs to Inhibit Virus-ACE2 Interactions
Much research has gone into finding drugs that can effectively prevent or reduce viral-ACE2 interactions. Drug repurposing has come to be an important strategy in this respect, as it can cut down the amount of time and the expense involved in developing a new drug. However, emergency drugs to treat COVID-19 remain scarce.
One such avenue of research is to find small molecules that can attach to residues at the site of binding of the RBD to the ACE2. The current paper reports the discovery of several potential binding sites for small molecules that can occupy such druggable pockets so as to inhibit virus-ACE2 binding in vitro.
The electronic microscopy model of the S protein was used to identify 6 possibly druggable pockets where drugs might bind to the interface region. Next, a genetic algorithm, in conjunction with local research, was used to virtually screen the molecule for binding by each of over 2900 FDA-approved drugs.
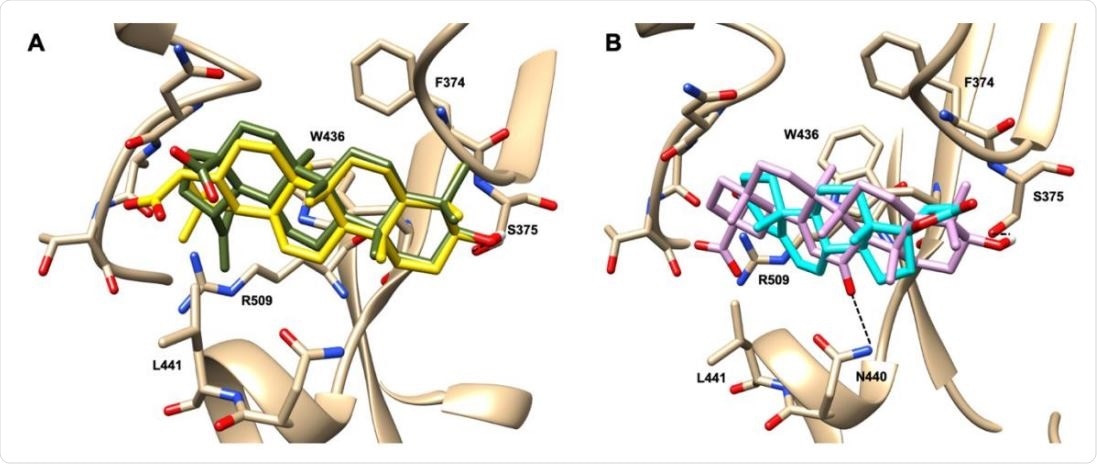
Graphical representation of the binding mode of the best compounds resulting from the screening in pocket 1. The RBD region is represented in transparent surface colored by residues hydrophobicity. Color codes are: dodger blue for the most hydrophilic regions, white, to orange-red for the most hydrophobic. (A) Betulinic acid (dark olive-green stick) and oleanolic acid (gold stick). (B) Glycyrrhetinic acid (plum stick) and potassium canrenoate (cyan stick). For clarity reasons hydrogen atoms are omitted and only interacting amino acids are displayed in sticks.
This led to the identification of several steroidal and triterpenoidal compounds that bind robustly and selectively to the RBD pocket 1 and pocket 5. These include glycyrrhetinic acid, betulinic acid and betulin, canrenone and potassium canrenoate, spironolactone, and oleanolic acid.
The best-performing compound was glycyrrhetinic acid, which has hydrophobic and hydrophilic interactions with the pocket. Similar modes of binding were shown by oleanolic acid and betulinic acid, while potassium canrenoate had a different binding orientation.
Inhibitory Natural or Semisynthetic Bile Acids
The researchers also looked at whether other compounds that bind other bile acid-activated receptors, namely, the Farnesoid-X-Receptor (FXR) and G protein Bile Acid Receptor (GPBAR)-1, could bind the above binding pocket of the RBD. These include oleanolic, betulinic and ursolic acid, and glycyrrhetinic acid and glycyrrhizic acid, which bind to these bile acid receptors. Bile acids are steroidal molecules that are produced in the liver.
They then looked at natural and semi-synthetic bile acids that are either now being used in medical treatment or are in the preclinical and clinical stages of development. These were tested for their binding ability with respect to the RBD binding pocket. These were found to have a higher affinity for pocket 5, which is very conserved.
Docking studies show that compounds with flat steroidal scaffolds and those with a cis A/B configuration dock to pocket 1 and pocket 5, respectively. These pockets have been confirmed to be functionally relevant to RBD-ACE2 binding by in vitro testing. Thus, effective inhibition of this binding is produced by several steroidal molecules (betulinic acid, glycyrrhetinic acid, oleanolic acid and potassium canrenoate).
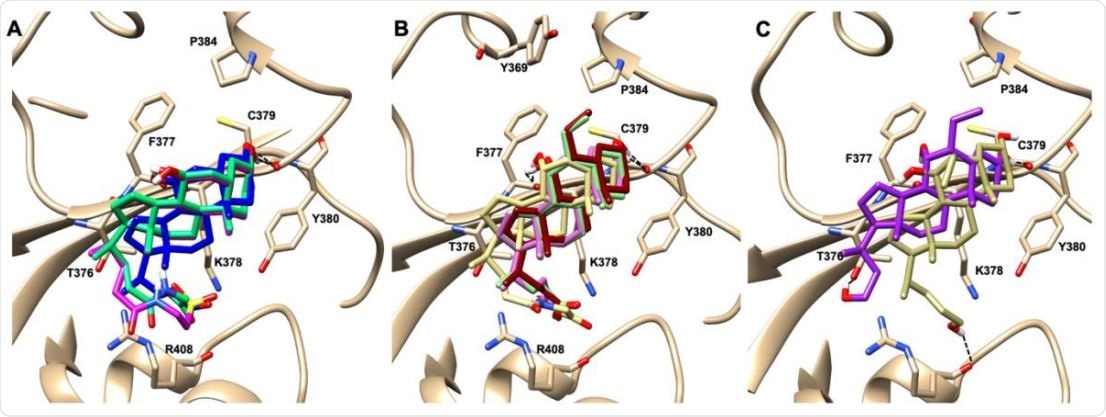
Graphical representation of the binding mode of the best compounds resulting from the screening in pocket 5. The RBD region is represented in tan cartoon, while the pocket 5 residues as transparent surface colored by residues hydrophobicity. Color codes are: dodger blue for the most hydrophilic regions, white, to orange-red for the most hydrophobic. (A) UDCA (blue stick), T-UDCA (magenta stick) and G-UDCA (spring-green stick); (B) CDCA (orchid stick), OCA (light-green stick), BAR704 (dark-red stick) and G-CDCA (khaki stick); (C) BAR501 (gold stick) and BAR502 (purple stick). For clarity reasons hydrogen atoms are omitted and only interacting aminoacids are displayed in sticks.
Structural Determinants of Inhibitory Activity
In particular, glycyrrhetinic and oleanolic acid showed very promising results. Oleanolic acid, which had only hydrophobic interactions with the pocket, produced the greatest inhibition among the screened drugs, suggesting that hydrophobicity is the main factor deciding inhibition efficiency.
This is also true of bile acids, both natural and semi-synthetic, which also showed the important role played by the 6α-ethyl group for inhibition. This group might promote the bioactive or ‘up’ conformation of the RBD, as well as facilitating hydrophobic contacts with other residues.
The vital determinant is, therefore, the network of hydrophobic contacts and not hydrogen bonding. Again, the polar group on the side chain appears to be non-essential for inhibition.
Implications and Future Research
Of all the natural and semi-synthetic agents tested, glyco-ursodeoxycholic acid (G-UDCA) had robust inhibitory activity (50%) against the RBD-ACE2. The semi-synthetic derivative BAR704, a highly selective and potent FXR agonist, was also effective and reduced the binding by approximately 50% at the dose of 0.1 and 1μM.
However, most of them were promoters at the two primary bile acid-activated receptors. In other words, all of these drugs bind to both these receptors and also to the viral protein. This finding could be used to hijack the virus-ACE2 binding. Combinations of these drugs did not increase the inhibitory efficiency over the use of single agents.
The researchers note that the best natural bile acid tested, G-UDCA, had a markedly less inhibitory activity than neutralizing antibodies from hyperimmune recovered COVID-19 patients, which achieved almost 99% inhibition. When the viral load is very high, therefore, small molecules may be almost useless, but could perhaps help prevent infection if only very mild exposure has occurred.
The main importance, therefore, lies in the indication of future directions of research, whereby these binding modes could be optimized by a better understanding of the steroidal structure. This could help evolve more interactions in pocket 5, which is the most conserved among currently circulating viral strains.
The investigators say, “The two pockets we have identified in β-sheet core of the Spike RBD appears to be targetable by steroidal molecules and importantly we have found that either naturally occurring bile acids and their metabolites in human seem be able to reduce the binding of Spire RBD with ACE2. Together, these results might help to define novel treatments to reduce the viral load by using SARS-CoV-2 entry inhibitors.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources