A new study by researchers from the United Kingdom and Taiwan and published on the preprint server bioRxiv* in June 2020 describes the structural basis for the neutralizing effect of the SARS-CoV-2 antibody on the virus. This also indicates a possible therapeutic molecule by identifying its binding epitope as possibly a major neutralizing site.
With the high mortality of the virus in certain subgroups of the population, the need for a therapeutic intervention other than supportive intensive care has become obvious. One such option which looks attractive against the current background is convalescent human plasma, which contains specific antibodies against the virus.
The promising feature of immune serum is its potential for the treatment of COVID-19 even at relatively late stages. For this to become more widely used, it is necessary to identify the combination of antibodies that uses different mechanisms to minimize the possibility of the virus evading the host immune defenses, while also preventing antibody-dependent enhancement.
The Spike Protein
The spike protein of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a type I transmembrane glycoprotein that is composed of S1 and S2 subunits. It has a cleavage site between the two subunits, and following binding to the ACE2 receptor or treatment with trypsin, it is cleaved into the two subunits at the N-terminal, and C-terminal ends respectively.
SARS-CoV-2 viruses binding to ACE-2 receptors on a human cell, the initial stage of COVID-19 infection, conceptual 3D illustration. Credit: Kateryna Kon / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Prior to fusion, the receptor-binding domain (RBD) at the apex is folded down like a hinge or flap, covered by the N-terminal domain of the spike. This renders its receptor binding site hidden unless the RBD swings up. At this point, it is able to bind the ACE2 binding site.
This interaction between the RBD and ACE2 locks it in the ‘up’ conformation, triggering the conversion to a post-fusion conformation. In this state, the S2 subunit attaches to the host membrane and sheds the S1 subunit.
Cross-reacting Antibodies to SARS-CoV-2
Antibodies that neutralize the ACE2 binding for SARS-CoV are typically not effective against SARS-CoV-2 and are rendered ineffective by escape mutations, such as the natural mutation Y495N at this site. On the other hand, the CR3022 antibody, from the blood of a SARS-CoV patient, does cross-react strongly with SARS-CoV-2 and also recognizes another hidden but highly conserved epitope on the RBD which is different from the binding site for ACE2. This is the EY96A epitope.
The researchers found that EY6A, one of the antibodies cloned from the plasma cells in the blood of a convalescent COVID-19 patient, binds the S1 subunit of SARS-CoV-2 as well as SARS-CoV. It was shown to have a high affinity for the RBD of the former virus. Both EY6A and CR3022 had interdependent binding, though asymmetrically.
EY6A vs. CR3022
In other words, if the RBD is expressed on the target cell, it was bound by ACE2 in the presence of EY6A; but if ACE2 is expressed on the cell, EY6A inhibited the RBD-ACE2 interaction. Measured this way, EY6A is about seven times stronger at blocking ACE2 binding than CR3022. It blocks ACE2 as well as it does ACE2-Fc and the molecule VHH72-Fc.
The researchers suggest that this means EY6A produces an indirect effect after binding to the RBD, which could be an allosteric or even a weak direct inhibitory interaction. When tested against CR3022, the latter is found to bind at a site far away from that which binds to ACE2, but in a completely competitive manner. This could mean they bind to the same or to very close or overlapping epitopes, but with tighter binding for the EY6A.
Neutralization tests using wild-type SARS-CoV-2 showed 1) that EY6A reduced the viral signal on qPCR testing by about a thousand-fold, confirmed by 2) a plaque reduction test.
Crystal Structure of EY6A-RBD Complex
The researchers determined the crystallographic structure of the SARS-CoV-2-RBD complexed with EY6A-Fab, either the latter alone or in complex with a nanobody that competes with ACE2. They found that the nanobody did not interact with EY6A, but that the nanobody bound to an epitope near and slightly overlapping the ACE2 binding site, on the RBD, at right angles to the EY6A.
As predicted, the EY6A binds to the same epitope that binds CR3022 but at a different orientation, at about 73 degrees to the perpendicular axis of the alpha3 helix of the RBD, which is the central helix for both epitopes. The binding is mediated via multiple hydrogen bonds and a salt bridge.
The RBD-EY6A binding triggers a conformational change at the alpha2 and alpha3 helices like those that occur with CR3022 binding. Thus, these three epitopes (EY6A, CR3022, and VHH72) have a high degree of overlap but with widely different orientations, so that VHH72 inhibits ACE2 binding.
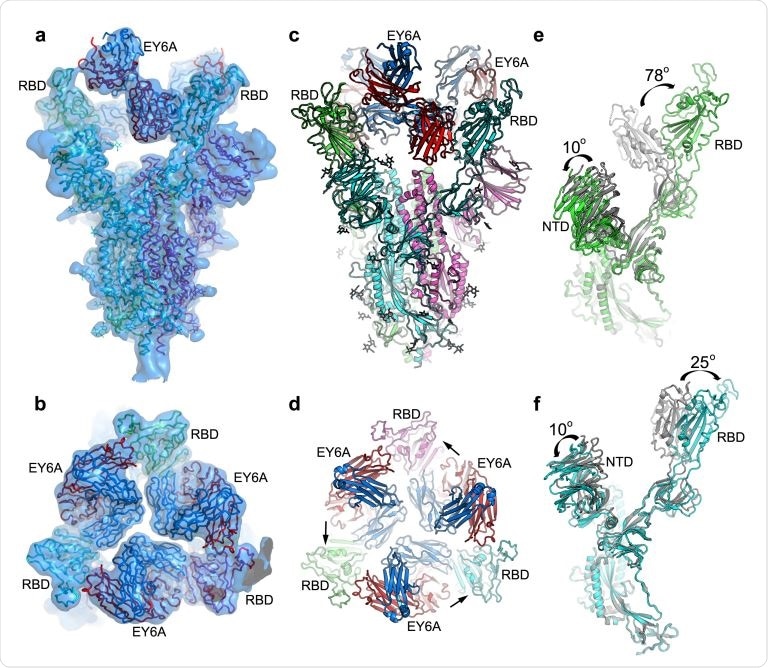
EM structure of SARS-CoV-2 spike and EY6A Fab complex. a & b, Side (a) and top (b) views of the cryo-EM density map drawn as semi-transparent surface showing three EY6A fabs bound to the Spike. c & d, Side and top views of the overall structure of the Spike-EY6A complex. Chains A, B and C of the Spike trimer and heavy and light chains of EY6A are shown in green, cyan, magenta, red and blue, respectively. The arrows in (d) indicate which RBD each Fab is binding. e & f, Comparison of the EY6A bound Spike structure with a reported open form Spike structure (grey; PDB ID 6VYB) with A chain in ‘down’ conformation (e) and B chain in ‘up’ conformation (f). NTD, N-terminal domain
Mutation Preventing Post-Fusion Change
Prior to fusion, the two residues in the linker between two helices in S2 were mutated to Proline-Proline, in order to avert the conformational change to the post-fusion state. In this structure, the RBDs were either all in the ‘down’ state or one ‘up’ and two ‘down’, with the EY6A being unexposed in either case. This mutation on the S2 protrusion causes the epitope to be pressed against it, in a buried protein-protein interface and completely hidden. If in the ‘up’ configuration, at least one more RBD must also be ‘up’ to open the epitope to the antibody.
Cryo-electron microscopy showed that the central axis at the top of the spike protein had three EY6A Fabs bound around it, with all the RBDs being in the ‘up’ state and rotated outside by 25 degrees. The spike is left extremely vulnerable to being broken up and requires the Fabs to stabilize it
Neutralization Mechanisms
An important mechanism of neutralization is blocking the virus attachment to the receptor. The current study suggests that another mechanism is the attachment of the virus to the EY6A epitope, which has been described by several researchers as binding to multiple antibodies and nanobodies targeting SARS-CoV, MERS-CoV, and SARS-CoV-2. A third possibility is the CR3022 antibody, which synergizes with ACE2 blocking antibodies to neutralize the virus.
Though the ACE2 receptor is distant in space from the EY6A, binding of the virus at these sites does produce some crosstalk between them.
The EY6A epitope is very unusual in that it is completely hidden in the Spike protein prior to fusion. The mechanism is explained as follows. Before fusion, the EY6A/CR3122 epitope rests on the upper end of the helix-turn-helix between the heptad repeat 1 and the central helix of S2 subunit like a lid. This prevents the spring-loaded helix extension that occurs after fusion, as a result of the mutation that is meant to suppress the post-fusion conformational change.
Implications
The EY6A residues are highly conserved because they are essential for these interactions, which is why mutations that avoid antibody binding have never been found. The epitope binds tightly to the RBD in isolation, about an order of magnitude more tightly than CR3022.
The position, on top of the Spike protein, where the EY6A binds, allows three Fabs to bind to the central axis at the same time, but not the CR3022 Fab. Accordingly, two-thirds of the spike molecules incubated with this epitope for 5 hours remain in the pre-fusion non-infective state. This is quite probable with intact antibodies as well.
The researchers comment, “In general, we would expect binders at this epitope to neutralize by displacing the ‘lid’ on the HR1/CH turn.”
This would destabilize the pre-fusion state and promote its conversion to the stable post-fusion spike trimer. However, the proline mutations introduced at the turn between successive helices avoid this conversion. Too-early conversion renders it unable to bind to the cell, and therefore it loses infectivity. The speed at which this process occurs decides how effective the antibody will be at neutralizing the virus, and thus in protecting against infection.
Another possibility is that the RBD being a small domain could allow different epitopes to interact. In fact, EY6A could have allosteric effects with ACE2 binding. In the same way, VHH-72 is capable of binding an epitope that overlaps with EY6A and is a potent inhibitor of ACE2 binding because it uses a different pose.
The study sums up: “Attachment to this single epitope can cause neutralization via more than one mechanism, and can exhibit strong synergy with ACE2 blocking antibodies.”
Moreover, with key amino acids being highly conserved, antibodies that tightly bind to this epitope might also be capable of neutralizing many other similar viruses such as SARS-CoV and perhaps MERS-CoV. This epitope should be the foundation of many novel therapeutics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources