The ability of coronaviruses to rapidly evolve, adapt, and cross species barriers make the development of effective and durable therapeutic strategies a challenging and urgent need. As for other RNA viruses, genomic RNA structures are expected to play crucial roles in several steps of the coronavirus replication cycle. Despite this, only a handful of functionally conserved structural elements within coronavirus RNA genomes have been identified to date. Now, in new research published on the preprint server bioRxiv* European scientists have performed RNA structure probing to obtain a single-base resolution secondary structure map of the full SARS-CoV-2 coronavirus genome. Their aim is to lay the foundation for the development of innovative RNA-targeted therapeutic strategies to fight SARS-related infections.
RNA viruses have the information needed to regulate the host cell for their benefit, stored at two levels. The first is the linear RNA sequence, encoding the various proteins they need to hijack the host cell operation and build new virions for replication and transmission. The second is the secondary and tertiary structure of the RNA genome provided by the intricate folds. These are necessary for the virus to replicate itself, synthesize proteins and package them, express molecules that help to avert an immune attack, and other functions.
RNA Viruses Have High Mutation Rates
RNA viruses associated with many deadly diseases have higher mutation rates than DNA viruses, that allow them to evolve quickly and to adapt to changing conditions, such as exposure to vaccines and drugs, simply by small alterations of the essential proteins. At the same time, they conserve certain RNA structures in the viral genome to a high degree, even with changes in the sequence and, therefore, of the amino acid. This makes these conserved structures important targets for therapeutic development.
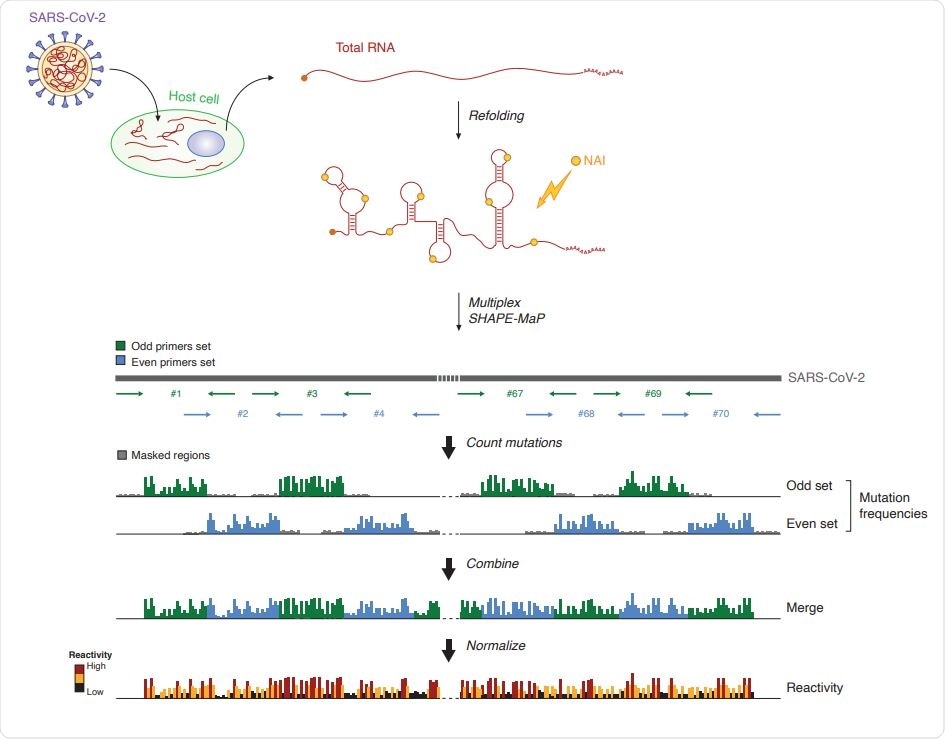
Genome-wide SHAPE-MaP analysis of SARS-CoV-2. (A) Schematic of the multiplex targeted SHAPE-MaP approach for querying the SARS-CoV-2 genome from total RNA.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Coronaviruses and Drug Targets
Coronaviruses (CoV) are RNA viruses, formerly thought to be non-pathogenic but now displayed as human pathogens, in the wake of the SARS, MERS, and the ongoing COVID-19 outbreaks. The ability of this viral genus to survive within a variety of reservoirs, to jump across species, and to cause high levels of sickness and death in humans, makes it a source of repeated human outbreaks. In fact, bat CoV sometimes can infect human cells as they are, without any further change, which means that these represent constant threats to public health.
Scientists have been trying their best to identify drug targets within the virus to bring down the mortality and the spread of this virus. One such promising target is the RNA structure, which indeed has already been shown to be capable of inhibition by small molecules directed against it in other viruses like HIV, HCV, and influenza A.
Identifying Structural RNA Elements
CoV have the largest genomes among all RNA viruses at approximately 30 kb. Earlier research has identified some structural cis-regulatory RNA structural elements using phylogenetic analysis. Among these are structures in the 5’ and 3’ untranslated regions (UTR) and the ribosomal frameshifting element (FSE).
The first type in most betaCoV includes stem-loops, SL 1-5, needed for viral replication. The FSE is at the boundary of ORF1a-ORF1b and is required to translate the ORF1a protein. Another similar pseudoknot to this FSE is thought to be at the 3’ UTR, with one highly conserved 8-nucleotide segment inside the hypervariable region and an SL-like 2 motif.
With more than 80% homology, investigators predict the presence of these RNA structural elements in SARS-CoV-2 as well, but this is not established as yet due to the difficulty of elucidating the structure of this large genome. This current study aims at characterizing the full-length RNA genome of a CoV, based on the SARS-CoV-2. The first step is to generate a model of the whole genome. The next is to find those regions that do not fold easily so as to use them to prepare antisense oligonucleotides to counter the infection. They also found a set of RNA structures with stable folds. Among these, about a tenth are subject to selective pressure.
Predicting The 3D Structure of RNA Elements
Using the secondary structure parameters with 2D modeling, they predicted the 3D structure of the elements so as to find which would best interact with small inhibitory molecules. This work is thus a “cornerstone”, according to the researchers, in the development of RNA-targeting strategies against SARS and similar infections.
They used an in vitro refolding technique of the total RNA from cultured cells infected with the SARS-CoV-2 so that the post-transcriptional modifications that might occur in vivo with natural infection, and could affect folding, are preserved, and high amounts of viral RNA can be analyzed.
The SHAPE-Map profiling of RNA structure showed that over half of the viral genome is structured. The known RNA structural elements typical of betaCoV were all identified, showing the accuracy of the model. However, some differences were also found, such as a single large SL-like structure rather than three SL (SL8 to SL 10).
Using Shannon entropy measurements along with SHAPE data, they differentiated genomic regions into those that were stably folded into easily recognizable conformations and those which stayed single-stranded. The latter is ideal for generating antisense oligonucleotides as they will typically not form any structure or take part in base pairing within the molecule.
Implications of RNA Structure on Function
They also examined the extent of conservation, and found about 24% of the single-stranded to be highly conserved, with over 80% conserved in SARS-CoV, and well over 60% for MERS and other betaCoV. Five of the regions were extremely conserved at above 86% for SARS-CoV, and 77% for MERS, and 74% for other betaCoV. This could indicate the presence of a key regulatory element in the RNA at these regions.
The easily folded regions were identified to have about 150 structure elements making up about 40% of the genome, including both the 5’ and 3’ UTRs. The researchers attribute RNA function to these regions in terms of mechanisms of attachment, entry, and viral replication and release, as well as structural aspects necessary for binding small and large molecules.
Using further steps to identify druggable targets, they found a pocket at the bottom of the 3′ stem-loop II-like motif (s2m), which is a highly conserved region found in other viruses as well. it could influence host translation via ribosomal protein interactions or after being processed to mature microRNA. The FSE was also found to have a perhaps druggable site, which is essential for viral fitness because it permits both structural and functional viral proteins to be synthesized. Within this FSE pocket is a smaller pocket that may be vulnerable to a small molecule.
Finally, they found 12 of the original 150 (approximately) RNA structural elements, which had significant covariates for at least 5% of the original base pairs. This could support the presence of functional RNA structures.
The study concludes, “Overall, our analyses reveal structures of the SARS-CoV-2 virus RNA that may turn out to be its weak spots. Not only our data will provide a fundamental resource for the development of innovative RNA-targeted therapeutic strategies, but also it will help to elucidate still unknown aspects of the life cycle of coronaviruses.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources