The causative agent of coronavirus disease (COVID-19) that, in some cases, leads to severe atypical pneumonia in humans, was swiftly identified as a novel coronavirus and designated as SARS-CoV-2.
Fundamental research that followed its discovery found that SARS-CoV-2 exploits the angiotensin-converting enzyme 2 (ACE2) receptor to enter host cells, which is primarily facilitated by viral spike protein.
Subunits of the aforementioned spike protein contain a receptor-binding domain (RBD) that can switch between an exposed 'up' conformation and a 'down' conformation, the latter being inaccessible for ACE2 binding.
.jpg)
SARS-CoV-2 viruses binding to ACE-2 receptors on a human cell, the initial stage of COVID-19 infection, conceptual 3D illustration. Image Credit: Kateryna Kon / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In any case, halting this binding event would stop the infection; hence, therapeutic neutralizing antibodies constitute a pivotal short-to-medium term approach to tackle the COVID-19 pandemic. Nonetheless, a traditional approach to antibody production is hindered by costly production and protracted development times.
As an alternative, nanobodies (i.e., single-domain antibodies) can also be utilized for treatment purposes, with several advantages: their small size, production simplicity, and increased stability. Consequently, the development of libraries of sybodies enables a swift, cheap, and stringent selection of binders against therapeutic targets.
In this study, a research group from Germany, Sweden, South Africa, and Switzerland reported the selection, biophysical, and structural analysis of sybodies that may be used for treatment purposes.
Sorting through a library of sybodies
Synbody selection on biotinylated RBD (i.e., when biotin is covalently attached to a protein) has been carried out with the three sybody libraries (concave, loop, and convex) following established procedures. In this project, a total of 33 sybodies were derived from the concave, 32 from the loop, and 20 from the convex library.
The enrichment of binders against RBD was closely observed by quantitative polymerase chain reaction (qPCR). Moreover, to obtain an in-depth understanding of the neutralizing activity of the identified binders, the researchers performed bio-layer interferometry assays to monitor ACE2 binding to immobilized RBD.
In addition, to test whether the selected sybodies can effectively neutralize SARS-CoV-2, this group performed a neutralization assay with lentiviral particles pseudotyped with SARS-CoV-2 spike protein
Finally, to confirm all observations regarding neutralization properties, a competition test under equilibrium conditions was developed in order to appraise the binding of very potent sybody 23 (Sb23) to the spike protein. This was done in the presence or absence of ACE2 by using microscale thermophoresis.
Potent sybody 23 picked out as a frontrunner
The researchers have demonstrated that the combined approach used in this study represents a viable fast workflow for selecting binders with neutralizing activity against SARS-CoV-2, but also other newly emerging viruses. More specifically, it was shown that they could compete with binding to ACE-2 and effectively neutralize SARS-CoV-2 spike pseudovirus.
"In particular, Sb23 binds to recombinant RBD as well as to the pre-fusion spike glycoprotein with high affinity and shows very potent neutralization activity", say study authors in their bioRxiv paper. "A small-angle X-ray scattering model of an RBD-Sb23 complex indicated that Sb23 binds in the vicinity of the ACE2 binding site on the RBD", they add.
Finally, a cryo-electron microscopy structure of Sb23 bound to the spike shows that Sb23 is situated in the ACE2 binding site on the RBD in both 'up' and 'down' conformation – thereby efficaciously blocking ACE2 binding. Additionally, cryo-electron microscopy reconstruction unveiled a completely novel conformation of the spike, where two RBDs are in the 'up' ACE2-binding conformation.
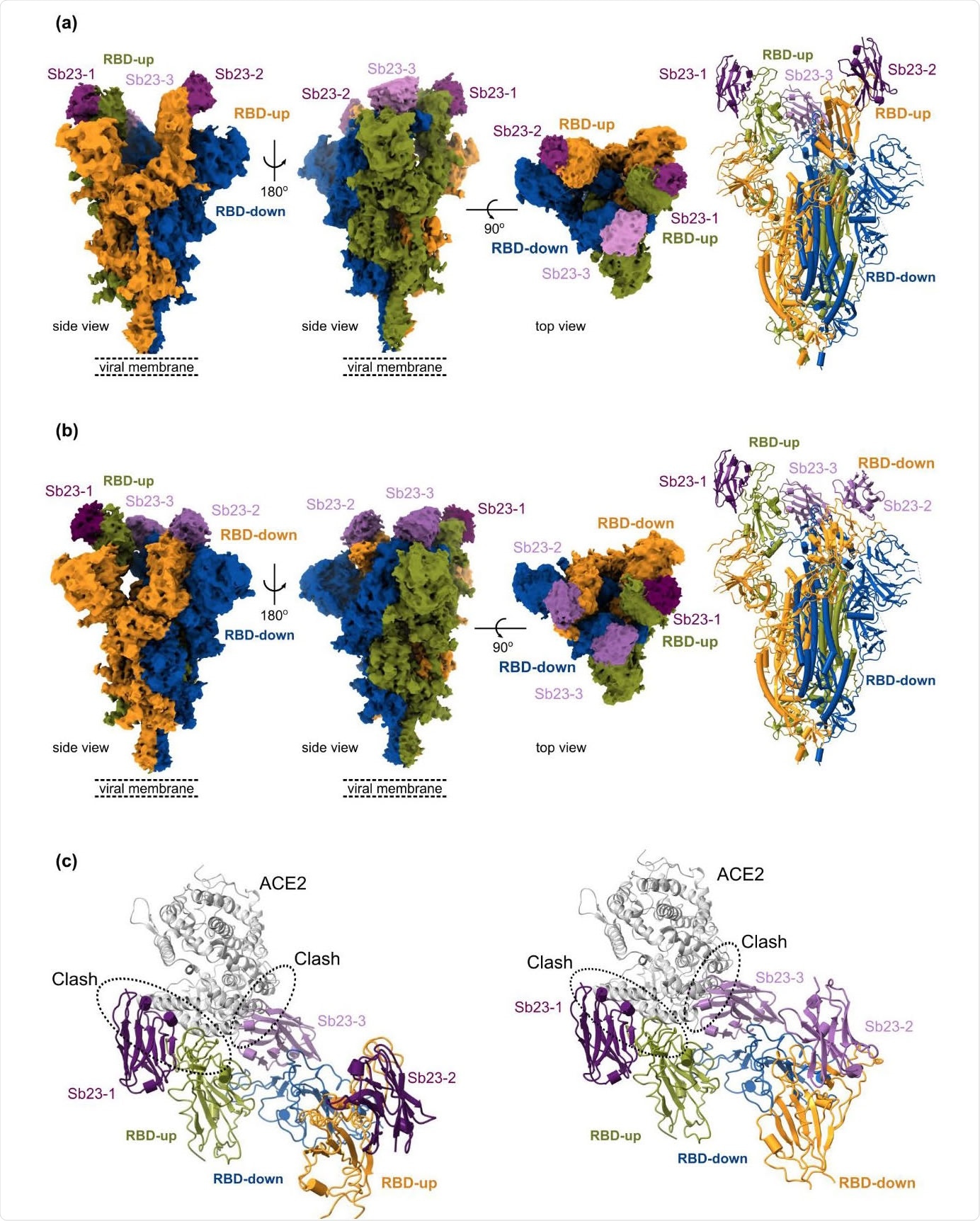
Cryo-EM reconstruction of SARS-CoV-2 spike bound to Sb23 - (a) Locally-sharpened Coulomb potential map and cartoon model of Sb23 bound to the spike protein in the ‘2-up’ conformation. (b) Locally-sharpened Coulomb potential map and cartoon model of Sb23 bound to the spike protein in the ‘1-up’ conformation. (c) Cartoon model of Sb23- bound Spike in the ‘2-up’ (left) ‘1-up’ (right) conformation showing how ACE2 binding is blocked by Sb23 bound to the RBD in the ‘up’ conformation as well as Sb23 bound to the neighboring RBD in the down conformation.
Effective binders as a potential therapeutic focus
Synthetic libraries are indeed a viable alternative approach to rapid drug development, with the promise of quickly generating highly specific binders with adequate neutralization potential. This inventive research endeavor indeed evidenced this.
None of the unique sybodies identified in this work were identical among two independent selections using the same libraries, underlining, in turn, the increased diversity of the three libraries, as well as their potential for choosing several different high-affinity binders.
Also, a novel conformation identified in this study makes previously unexposed epitopes accessible for the development of therapeutic binders within the central cavity of the spike, which includes the lower portion of the RBD and potentially also the central helical region of the spike protein.
In conclusion, the COVID-19 pandemic has opened our eyes to how previously unknown viral strains can rapidly emerge and spread across continents, causing significant casualties in a very short time frame. Therefore, the possibility of rapidly developing treatment against such novel viral strains is something we should nurture for the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Custódio, TF. et al. (2020). Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. bioRxiv. https://doi.org/10.1101/2020.06.23.165415.
- Peer reviewed and published scientific report.
Custódio, Tânia F., Hrishikesh Das, Daniel J. Sheward, Leo Hanke, Samuel Pazicky, Joanna Pieprzyk, Michèle Sorgenfrei, et al. 2020. “Selection, Biophysical and Structural Analysis of Synthetic Nanobodies That Effectively Neutralize SARS-CoV-2.” Nature Communications 11 (1): 5588. https://doi.org/10.1038/s41467-020-19204-y. https://www.nature.com/articles/s41467-020-19204-y.