SARS-CoV-2, 2019 nCoV virus diagram. Image Credit: Orpheus FX / Shutterstock
The new assay, called nanoluciferase SARS-CoV-2 (SARS-CoV-2-Nluc), generates neutralizing antibody results more quickly than the current gold-standard technique, shortening the turnaround time from three days to five hours.
It also performed reliable, high-throughput antiviral screening and identified the human rhinovirus drug rupintrivir, as a potentially effective inhibitor of SARS-CoV-2.
The authors say rupintrivir may now be considered for further testing to determine its potential as a treatment for coronavirus disease 2019 (COVID-19).
A pre-print version of the paper is available on the bioRxiv* server, while the article undergoes peer review.
Current testing techniques
Since the COVID-19 pandemic began in Wuhan, China, late last year, nucleic based RT-PCR (reverse transcriptase-polymerase chain reaction) testing has been used to diagnose people with acute viral infection and to trace and control contact transmission.
However, given the number of infected individuals who are asymptomatic, rapid, and reliable serodiagnosis is urgently needed to help establish the true scale of infection within communities.
Furthermore, being able to assess the level of neutralizing antibodies quickly would help researchers understand immunity post-infection, identify potential donors of protective antibodies, and assess vaccines that are currently in the pipeline.
Several serological assays have been developed, but the gold standard so far is the plaque reduction neutralization test (PRNT), since it directly determines the levels of neutralizing antibodies needed to inhibit wild-type virus.
However, the technique is low-throughput, with a turnaround time that makes it unsuitable for large-scale diagnosis of COVID-19.
The new assay is high-throughput
“A high-throughput platform would greatly facilitate COVID-19 serological testing and antiviral screening,” say the researchers.
Now, Pei-Yong Shi and colleagues have developed the SARS-CoV-2-Nluc platform, a reporter virus assay that exhibits similar replication to wild-type SARS-CoV-2 in vitro.
On using the assay to measure antibody neutralization in the sera of patients with COVID-19, the results generated were comparable to those of conventional PRNT.
Furthermore, the turnaround time achieved with the high-throughput SARS-CoV-2-Nluc assay (3 hours) was significantly shorter than is currently achieved with the low-throughput PRNT (three days).
Although the assay was performed in a 96-well format for this study, Shi and colleagues say the magnitude and dynamic range of the Nluc signal mean it could easily be adapted to a 384- or 1536-well format for large-scale testing.
“Thus, the assay can be readily deployed for large-scale vaccine evaluation and neutralizing antibody testing in humans,” write the researchers.
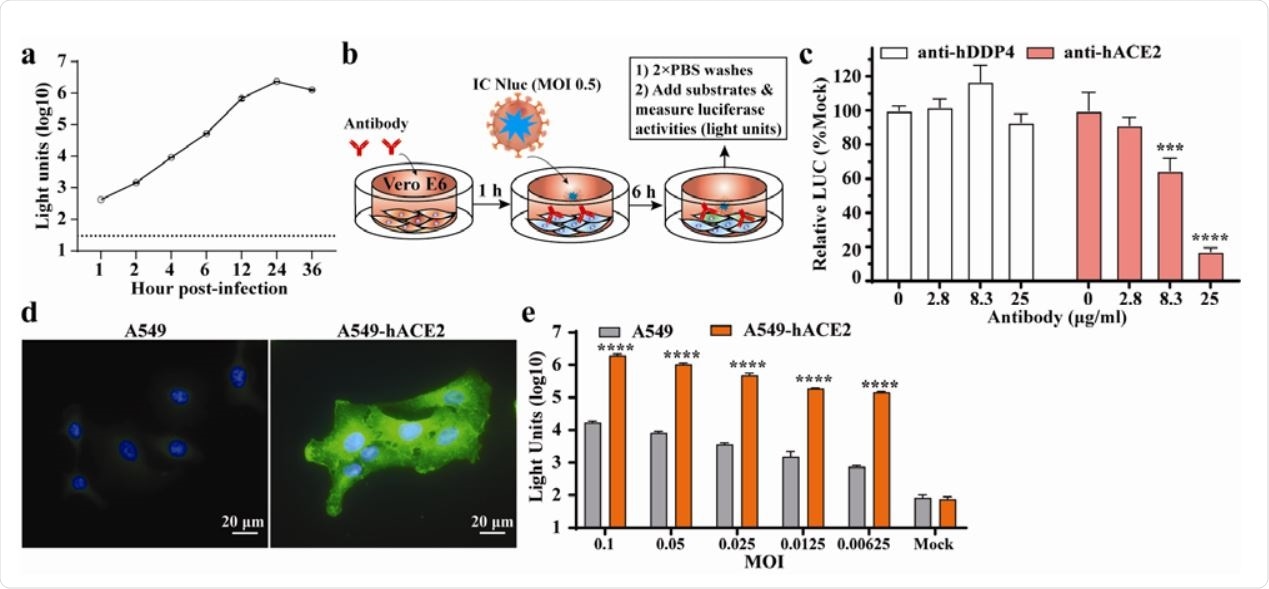
Application of SARS-CoV-2-Nluc in analyzing hACE2 as an entry receptor. (a) Replication kinetics of SARS-CoV-2-Nluc (IC Nluc) on Vero E6 cells. Cells were infected with IC Nluc at MOI 1.0. At given time points, cells were harvested for luciferase signal measurement. The means and standard deviations from three independent experiments are presented. (b) Diagram to analyze hACE2 for IC Nluc entry. (c) Relative luciferase signals following infection of cells that were preincubated with anti-hDPP4 or anti-hACE2 antibodies. The luciferase signals from antibody-treated groups were normalized to those from untreated groups. The average and standard deviation of three independent experiments are presented. (d) Immunofluorescence analysis of hACE2 expression in A549-hACE2 cells. At 24 h post-seeding, the cells were fixed and stained with anti-hACE2 polyclonal antibody. (e) Luciferase signals from IC Nluc infected561 A549 and A549-hACE2 cells. Cells were infected with indicated MOIs and luciferase signals were measured at 24 h post-infection.
Developing the assay for antiviral screening
Next, the team developed a version of the SARS-CoV-2-Nluc infection assay for the high-throughput screening of antivirals and validated the method using known inhibitors of SARS-CoV-2, including remdesivir and chloroquine.
They then used the assay to test various approved and investigational antiviral and anti-infective drugs, including ones known to work against HIV, hepatitis C virus, and human rhinovirus.
Of 40 compounds tested (including remdesivir and chloroquine used for validation), the assay identified the HIV drugs nelfinavir and cobicistat and the human rhinovirus drug rupintrivir as the most potent and selective inhibitors of SARS-CoV-2-Nluc.
“Based on their antiviral potencies established in vitro, it is unlikely that nelfinavir or cobicistat would exert major clinical effects in COVID-19 patients at the current clinically approved doses,” said Shi and team.
Rupintrivir may effectively inhibit SARS-CoV-2
However, rupintrivir is a selective covalent inhibitor of the viral 3-chymotrypsin-like cysteine protease (3CLpro) enzyme that regulates coronavirus replication, say the researchers.
Rupintrivir may, therefore, inhibit SARS-CoV-2 by blocking the main 3CLpro cysteine protease activity, they suggest.
“Further studies would be required to better understand rupintrivir’s mode of action, efficacy in animal models, 268 and potential clinical benefits in COVID-19 patients,” write Shi and colleagues.
A rapid, high-throughput, reliable tool
The researchers say that the SARS-CoV-2-Nluc assay represents a rapid, high-throughput, reliable tool for neutralization testing and antiviral drug screening.
“Since neutralizing titer is a key parameter to predict immunity, the rapid SARS-CoV-2-Nluc neutralization assay will enable many aspects of COVID-19 research, including epidemiological surveillance, vaccine development, and antiviral discovery,” they write.
Among the antiviral agents tested, “rupintrivir was identified as a selective in vitro inhibitor of SARS-CoV-2 that might be considered for further studies to fully establish its potential for the treatment of COVID-19,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources