The emergence of a novel coronavirus SARS-CoV-2 in China has quickly turned into a global pandemic of coronavirus disease (COVID-19). At the moment, there are no approved vaccines or targeted therapeutics for the treatment of this disease, while diagnostic tests are still striving to achieve optimal sensitivity and specificity.
On the other hand, fundamental structural studies of the virus are progressing at an unprecedented pace. It is already well-known that SARS-CoV-2 spike glycoprotein (S-protein) promotes cell entry and is the main target of serologic response, especially its receptor-binding domain (RBD); also, the virus uses angiotensin-converting enzyme 2 (ACE2) as a cell receptor.
.jpg)
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of an apoptotic cell (green) heavily infected with SARS-COV-2 virus particles (purple), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The promise of molecular probes
Molecular probes with the affinity for key targets of certain viruses can be of broad utility. They can serve as molecular tags to facilitate the identification of effective antibodies for passive therapy; additionally, they can be utilized to assess sera reactivity and to obtain sensitive markers of infection, but also to define and monitor elicited responses needed for vaccine development.
As a result, biotin-labeled molecular probes that comprise specific regions of the SARS-CoV-2 S-protein (i.e., a main viral target for neutralization antibody) may indeed be very helpful in the isolation and characterization of antibodies that target this newly emerged pathogen.
A research group from the US National Institute of Allergy and Infectious Diseases, Columbia University, Frederick National Laboratory for Cancer Research, and the University of Kansas described the structure-based design of molecular probes, encompassing SARS-CoV-2 spike and its domains.
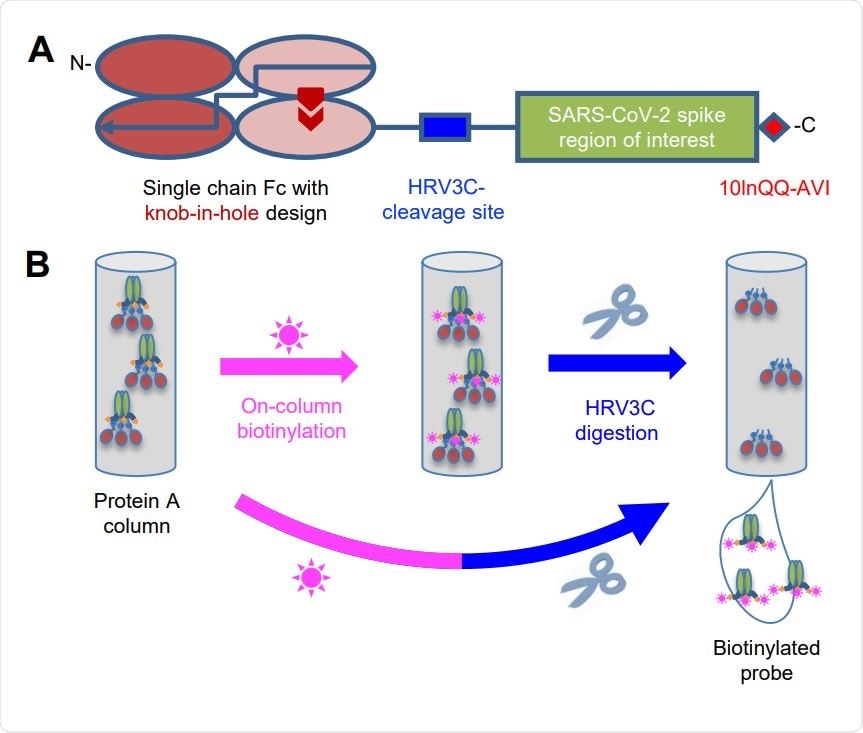
Strategy for Tag-Based Purification with On-Column Biotinylation (A) Schematic design of the expression construct of SARS-CoV-2 molecular probes. A single chain human Ig constant domain (scFc) was added at N terminus to facilitate expression and purification. The AVI tag was placed at the C terminus after a 10-amino acid linker for biotinylation. The red arrows in the second and fourth Fc domains showed the “knob-in-hole” mutations to prevent dimerization of the scFc. (B) Biotinylation and HRV3C digestion. Cell culture supernatant from cells transiently transfected with plasmid was loaded onto protein A affinity column. Biotinylation and HRV3C cleavage reactions can be carried out in series or simultaneously, as buffers for both reactions are compatible.
Stabilized SARS-CoV-2 spike trimer with biotinylation
For probe construction, the researchers used a process that incorporated 'cut-and-paste' assembly of obligatory components, employing an N-terminal purification tag and sequence stretches targeted by sequence-specific enzymes.
More specifically, the researchers first designed a construct that allowed for tag-based purification and on-column biotinylation (the latter being a process of covalently attaching biotin to a protein). Next, they incorporated the SARS-CoV-2 spike ectodomain with prefusion stabilizing mutations and a C-terminal trimerization motif.
All probes were characterized for antigenicity and ACE2 receptor recognition, and the structure of the spike ectodomain probe was additionally determined by cryo-electron microscopy. Antibody-binding specificities and cell-sorting capabilities of the biotinylated probes were also characterized in detail.
"We also used the structure of recombinant receptor binding domain with ACE2 to define mutations that could inhibit ACE2 interaction, which we incorporated into mutant RBD probes with ACE2-recognition ablated", study authors further explain their approach.
Finally, they have characterized additional properties of the devised probes – including the degree of biotinylation and potential use in sorting yeast cells expressing SARS-CoV-2 spike-binding antibodies or B-cells from COVID-19 convalescent donors.
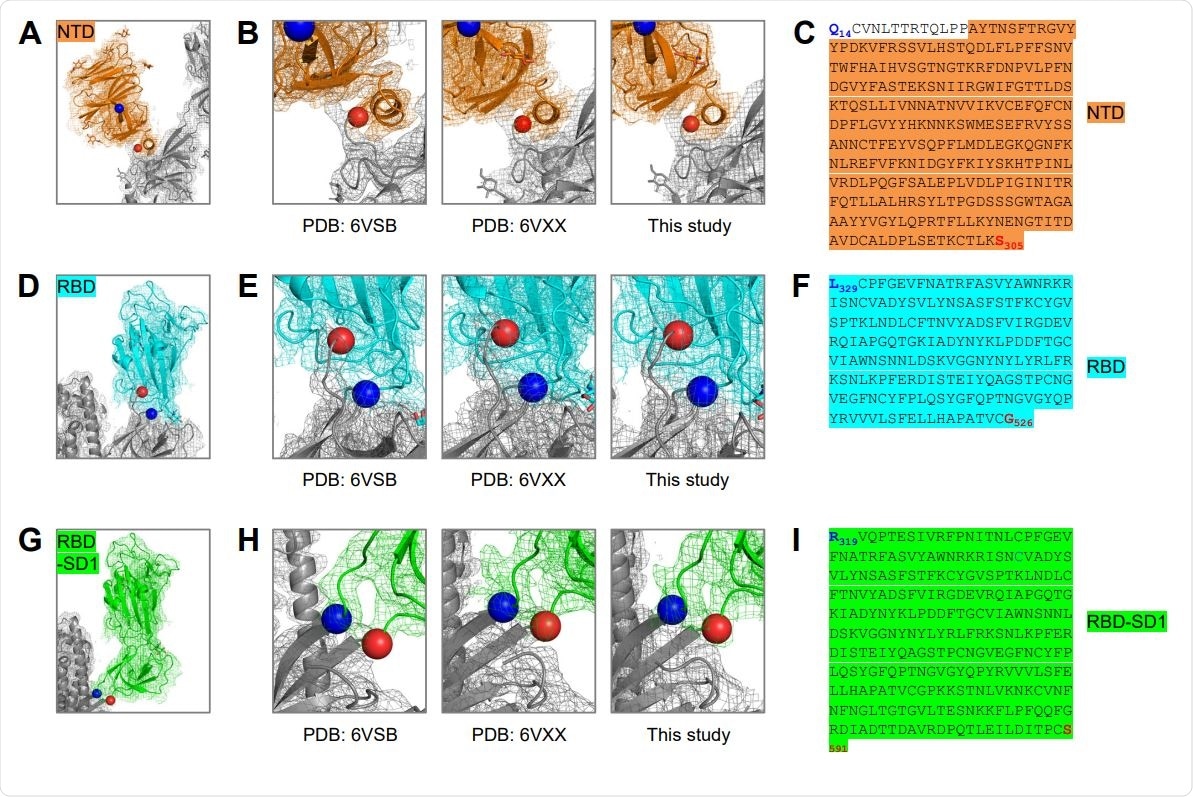
Structure-Based Definition of SARS-CoV-2 Molecular Probes Comprising the NTD, RBD and RBDSD1 Domains (A) Cryo-EM structure of the NTD domain in the S2P probe determined in this study (Figure 3E), with reconstruction density shown in orange for NTD domain, and gray otherwise. First ordered residue with density (A27) is highlighted with a blue sphere; last residue of NTD domain (S305) is highlighted with a red sphere. (B) Close-up view of the NTD termini. (C) Sequence of NTD domain probe. The sequence is highlighted in orange except for residues 14-26, which are disordered in the cryo-EM structures. (D) Cryo-EM structure of the RBD domain in spike (Figure 3E), with reconstruction density shown in cyan for RBD domain, and gray otherwise. First residue with density (L329) is highlighted with a blue sphere; last ordered residue of RBD domain (G526) is highlighted with a red sphere. (E) Close-up view of the spike RBD termini. (F) Sequence of RBD domain probe highlighted in cyan. (G) Cryo-EM structure of the RBD-SD1 domains in spike (Figure 3E), with reconstruction density shown in green for RBD-SD1 domain, and gray otherwise. First residue with density (R319) is highlighted with a blue sphere; last ordered residue of RBD-SD1 domain (S591) is highlighted with a red sphere. (H) Close-up view of the spike RBD-SD1 termini. (I) Sequence of the RBD-SD1 domain probe highlighted in green.
Towards the streamlined development of molecular probes
Altogether, this type of structure-based design (coupled to efficient purification and biotinylation processes) can enable streamlined development of SARS-CoV-2 spike-ectodomain probes. As demonstrated in this study, their properties hinted that such probes are actually a good biological mimic of the prefusion SARS-CoV-2 spike ectodomain.
"The structure-based methods we describe here for probe construction may allow for the assessment of immune responses of other type 1 fusion machines", say study authors. They also add that the truncated domain probes folded properly and preserved the native conformation.
"Overall, the cut-and-paste structure-based design described here should be easily adapted to the streamlined development of molecular probes against not only these pathogens, but also emerging pathogens, as shown here for SARS-CoV-2", conclude study authors.
Probe design can be considered an emerging field that has demonstrated its significance in informing further steps toward personalized medicine. As COVID-19 shows substantial individual variation in clinical presentation, individualized approach is increasingly important.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources