PrecisionLife’s unique AI platform has found 68 genes associated with risk of developing severe COVID-19 after analysis of the genomes of 929 patients from the UK Biobank who had a severe response to SARS-CoV-2.
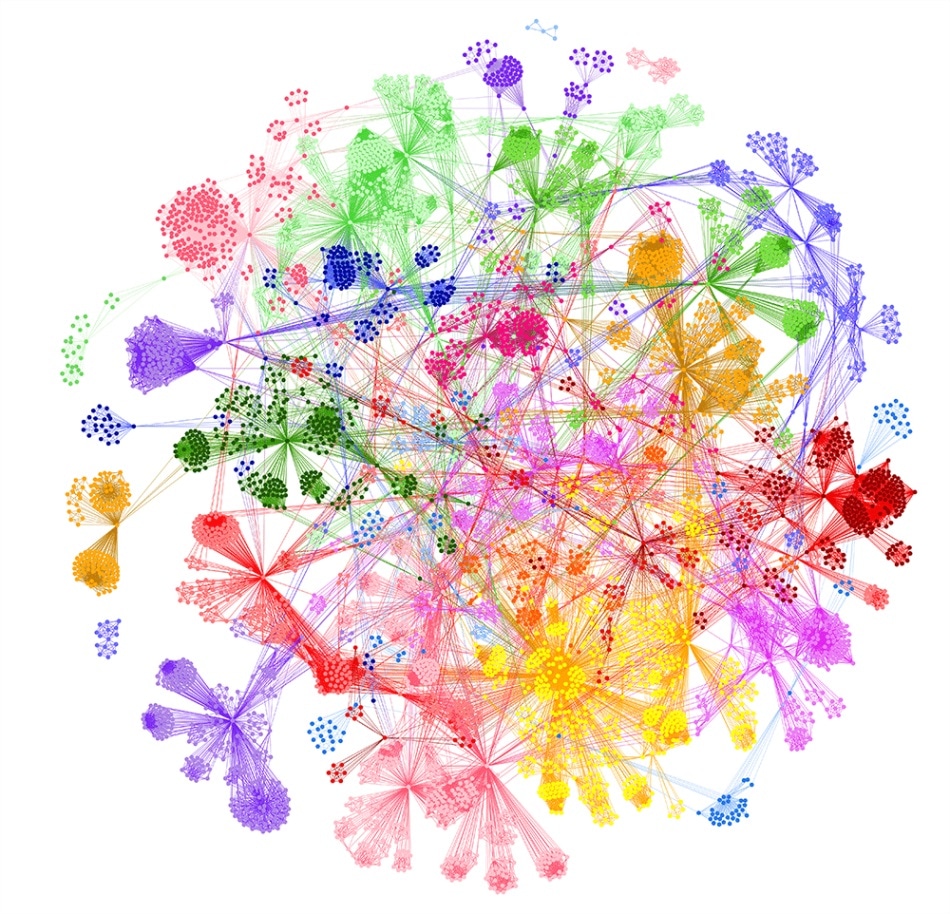
Disease architecture of the severe COVID-19 patient population generated by the PrecisionLife platform. Each circle represents a disease-associated SNP genotype, edges represent co-association in patients, and colours represent distinct patient sub-populations. Copyright PrecisionLife
Prior to an effective and widely available vaccine, PrecisionLife’s insights may help to identify patients who are at greatest risk of developing the most severe forms of COVID-19. They can inform the development of biomarker driven tests, targeted shielding and new therapeutic strategies, with the aim of identifying high-risk people, reducing disease burden and improving survival rates.
The results of this new study have been uploaded to the pre-publication site, medRxiv.
Established Genome Wide Association Studies (GWAS) approaches have been limited by the heterogenous nature of COVID-19, making it difficult to clearly explain the wide range of symptoms and impacts of predisposing comorbidities associated with the disease. The study has overcome this barrier by evaluating combinations of genetic features, which is not possible with existing GWAS approaches.
Using a combinatorial (high-order epistasis) analysis approach, PrecisionLife identified 68 protein-coding genes that were highly associated with severe COVID-19, nine of which have been previously linked to differential response to SARS-CoV-2. These 68 genes include several druggable protein targets and pathways, nine of which are targeted by drugs that have reached at least Phase I clinical trials.
This is our second study specifically looking at host genomics to identify those most at risk and find opportunities to treat patients with later stage severe disease where host immune processes begin to run out of control. We know that systemic remodeling, leaky vasculature and micro-clotting can cause severe issues in COVID-19 patients in lungs as well as other organs. We are encouraged that several of the genes identified relate to lipid programming, beta-catenin and protein kinase C signalling whose processes converge in a central pathway involved in plasma membrane repair, clotting and wound healing. This pathway is largely driven by calcium ion activation, which is a known serum biomarker associated with severe COVID-19 and ARDS. We intend to perform further analyses to investigate this hypothesis. We are also seeking to further validate 12 genes associated with dysfunctional immune response that are the first hint at a potential genetic signature for enhanced risk of flipping into a severe disease state.”
Dr Steve Gardner, CEO of PrecisionLife
Commenting on this encouraging data, Dr Duncan McHale, Co-founder of Weatherden, a clinical development consultancy said “Responses to infection with SARS CoV2 range from no symptoms at all to death with marked heterogeneity of clinical and pathological features. Significant associations of disease severity risk have been observed with epidemiological factors including age, sex, blood group and ethnicity in addition to co-morbidity with many common conditions such as cardiovascular disease, diabetes, hypertension and chronic pulmonary diseases including asthma. It would be extremely useful to be able to identify the underlying pathological features that result in differential host responses, particularly those that predispose some patients to developing severe COVID-19, enabling improvements in clinical care as well as the identification and development of novel therapeutic approaches.”
The Company has disclosed full details of the genes it has identified, including the druggable targets and pathways, to encourage rapid and collaborative use of the findings to improve the management of at-risk populations. As more genomic records and additional medical data become available in the UK Biobank and other data sources, PrecisionLife will extend its analyses to fully ascertain the differences between severe and mild COVID-19 cases and provide an additional layer of validation to the results from this study.
PrecisionLife notes that one limitation of the UK Biobank dataset is that the ethnicity distribution of the participants is heavily skewed to white British participants and it has consequently not been possible to fully investigate additional risk factors in BAME patients. It is actively seeking to investigate severe COVID-19 risk factors in other global datasets with more ethnically diverse populations. The addition of more phenotypic and clinical data in its analyses may also be used to gain greater understanding into the association of other observed epidemiological risk factors such as blood groups, environment, socioeconomic status and prescription medication history with development of severe COVID-19.