SARS-CoV-2 is a spherical particle with a single-stranded RNA genome, which has several regions – the genes encoding the replicase ORF1ab, spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins, respectively, from the 5’ end. The S protein facilitates viral entry by binding to the host receptor, the angiotensin binding enzyme 2 (ACE2), and has the highest variability among the genetic sequence, and is common to both SARS-CoV-2 and SARS-CoV. Still, the former has a much higher affinity for binding to the receptor, about 10-20 times greater.
.jpg)
SARS-CoV-2 viruses binding to ACE-2 receptors on a human cell, the initial stage of COVID-19 infection. Image Credit: Kateryna Kon / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Abundance of Genomic Data on SARS-CoV-2
Currently, there are many published genome sequences in various published databases - almost 54,000 in GISAID, about 24,000 in NCBI, and almost 8,000 in ViPR. Three major virus clades are observed that show non-uniform distribution over different geographical regions, probably because of founder mutations. Most Asian isolates are from superclade I, while US isolates are mostly in superclade II. Most European and US East Coast isolates are in superclade III.
Among the isolates of superclade III are many with a characteristic A23403G mutation, which is considered to confer increased viral fitness, perhaps since it is rapidly becoming predominant over other strains all around the world. It is also part of an antigenic epitope that induced antibody production to the earlier SARS-CoV virus. This may cause antibody escape and thus allow reinfection in those who have recovered from COVID-19.
The Study: Phylogenetic Analysis of the Virus in Eastern Ontario
The current study used 25 sequenced viral genomes from the earliest cases in Ontario, Canada, to recreate the phylogenetic descent of SARS-CoV-2. All the samples were from the earliest cases in this region, and therefore the source of infection was thought to be from travel-related contacts, directly or indirectly.
The genomes were all from viruses isolated from nasopharyngeal swabs, and the genome analysis was performed based on two main assumptions. First, they assumed that identical sequences were from the same ancestral strain, rather than evolving independently. Secondly, new mutations were assumed to arise at a rate of about 1 base pair every 7-21 days, as observed with the mean rate of about 24 bp substitutions per year in the GISAID’s Nexstrain analysis.
The individual genomes were queried against a local reference set of approximately 25,000 genomes retrieved from the GISAID database. They retained those genomes, which were remarkably similar, with two or fewer mismatches, thus narrowing down the number to about 1,200. When identical genomes from different subjects were matched, the final number came down to 72 unique sequences.
The root sequence was added, using a Wuhan sequence. The analysis showed that 59/72 sequences had the same common ancestry, with not more than a couple of substituted amino acids or other residues. Removing these different groups allowed the researchers to see only the genomes from the patients and valuable ancestral sequences.
Three Major Clades, Different Origins
The study showed that 25/45 variants found in all the genomes were in the form of substitutions of cysteine to thymine, and then guanine to thymine in 7 variants. All the shared variants were present on both alleles. The researchers found that there were two large clades, two with four mutations in the S clade (C8782T and T28144C) and 23 (C241T, C3037T, C14408T, and A23403G) with four mutations in the G clade. The latter has smaller clusters.
Phylogenetic analysis of the local genomes was then matched with the reference sequences, which showed the 13 ancestral genomes that underlie the origins and the spread of these Canadian genomes. This includes the ancestral genome seen with reference 1, and now seen almost only in North America, mostly in the USA.
Another set of samples belongs to cluster 1 of the G-clade, mostly in Europe, and particularly in the UK and Spain. Patients with these strains have a history of European travel, which confirms this trail.
Several other samples from G-clade cluster 2 are from a few ancestral sequences found mostly in North America, and here too, the travel history supports the American origin of the infection in these cases. There were four clusters of samples, within G-clade cluster 2, which have the same genome, and were probably infected from the same source.
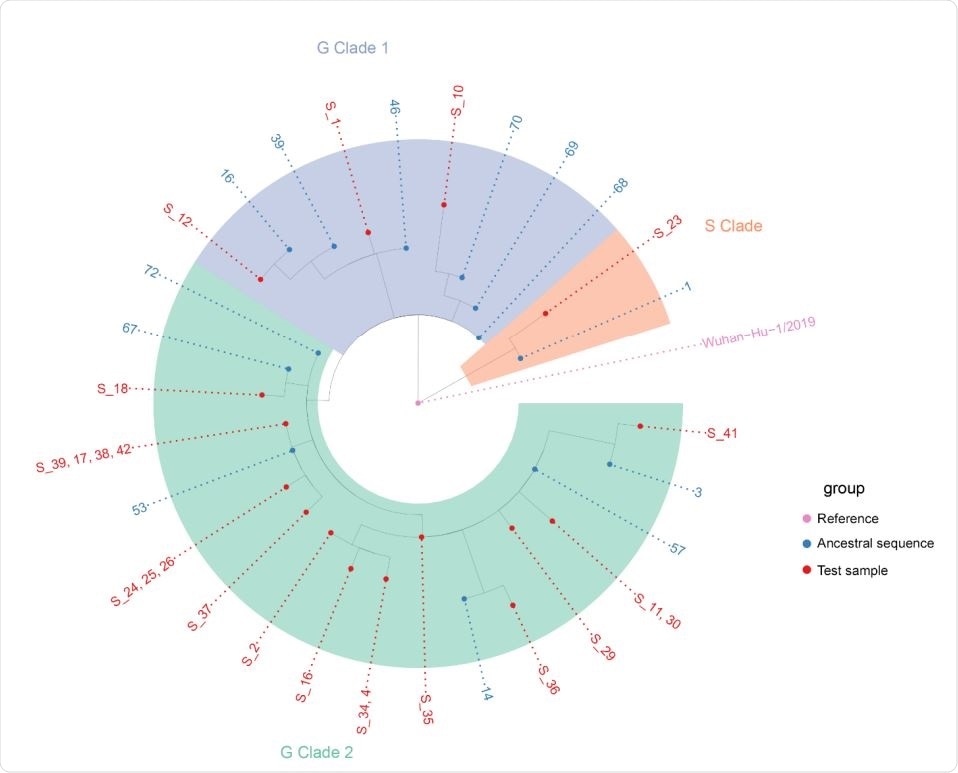
Nextstrain phylogenetic tree of local cases of SARS-CoV-2 and the most similar ancestral sequences in the GISAID database. Sample 23 in the S-clade appears derived from ancestral sequences 1 which predominately includes sequences from North America (USA). Samples 1,10, and 12 in the G-clade 1 is derived from ancestral sequence 68 and others which predominately appear European (UK, Spain, Portugal) in origin. The other 19 samples in the G-clade 2 is derived from ancestral sequence 72 and others which appear predominantly North American (USA) in origin.
The S protein of SARS-CoV-2 is considered to limit the host range and is also the immunodominant antigen. The current analysis succeeded in finding three unique and new mutations in the S region, all of which are synonymous and will, therefore, conserve both virulence and epitopes. They also found another site with a missense variant, C25217T, with glycine to cysteine substitution, but its effect is still to be understood.
There were five heterozygous variants, four found only in one sample, and one shared between two samples.
Overall, the researchers noted, “These correlations between phylogenetic origin and reported travel history indicates how viral genome sequencing can successfully trace the origin of SARS-CoV-2 infections into Canada.” The samples that showed identical sequences might have been either from a common source of infection or because of community transfer. The latter is likely because these patients have no recorded history of travel outside Canada but had contact with each other.
Puzzling Facts from Phylogenetic Analysis
There are other samples with the same sequence but where there is no known connection between the individuals. It is important to find the trace among all these patients so as to identify the major routes of transmission.
Other puzzles are also present, such as identical samples from two patients of which one was positive five days earlier. Still, the other sample had a history of US travel, and they have no known connection! This may mean that the later sample was not due to US travel after all.
Implications
Despite the limitations of the study, such as the lack of data on asymptomatic carriers, and the discrepancies in data collection which lead to incomplete travel and contact histories, it remains the earliest discussion of viral genomes related to COVID-19 from this part of Canada, including many of the earliest cases there.
The study raises questions as to the adequacy of the traditional explanations of infection, such as contact with travelers from foreign places with a higher burden of infection, or self-travel. While this may be true, the study suggests that community transfer may have already been occurring early in the reported course of the outbreak.
The study concludes, “These results demonstrate how molecular epidemiology and evolutionary phylogenetics can help local health units to track origins and vectors of spread for emerging diseases like SARS-CoV-2. Earlier detection and screening and alternative modes for contact tracing may improve the effectiveness of regional public health interventions to prevent future pandemics.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sjaarda, C. P. et al. (2020). Chasing the Origin Of SARS-Cov-2 In Canada’s COVID-19 Cases: A Genomics Study. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.25.171744. http://biorxiv.org/cgi/content/short/2020.06.25.171744
- Peer reviewed and published scientific report.
Sjaarda, Calvin P., Nazneen Rustom, Gerald A. Evans, David Huang, Santiago Perez-Patrigeon, Melissa L. Hudson, Henry Wong, et al. 2021. “Phylogenomics Reveals Viral Sources, Transmission, and Potential Superinfection in Early-Stage COVID-19 Patients in Ontario, Canada.” Scientific Reports 11 (1). https://doi.org/10.1038/s41598-021-83355-1. https://www.nature.com/articles/s41598-021-83355-1