The COVID-19 outbreak that began in Wuhan, China, has now spread all over the world, infecting over 10 million people and causing over 500,000 deaths. The biology and spread of the virus have been under intensive scientific scrutiny since then, given the urgent need to arrest the propagation of the virus with an effective vaccine or antiviral.
Human-Virus Matching
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the coronavirus (CoV) family that is found in many host species. The earlier severe acute respiratory syndrome (SARS) virus was studied to understand the route of spread and the potential animal reservoirs for such pathogens.
By the start of 2007, scientists had come to the view that bats harbored many novel viruses capable of crossing species barriers to infect human beings as well, but also many viruses closely related to the SARS-CoV. These viruses were known to be highly variable, which accounted for the greater risk they pose to humans and other domestic animals when compared to other viruses
The risk for zoonotic transmission increases the risk for such disease to emerge when there is frequent trade in wildlife globally. The underlying assumption is that if a virus is capable of replicating in multiple hosts, it adapts in a trade-off between precise and functional matching so that it can fit the wide range of tRNA molecules in different hosts. On the other hand, a single host will require specialized viral genes.
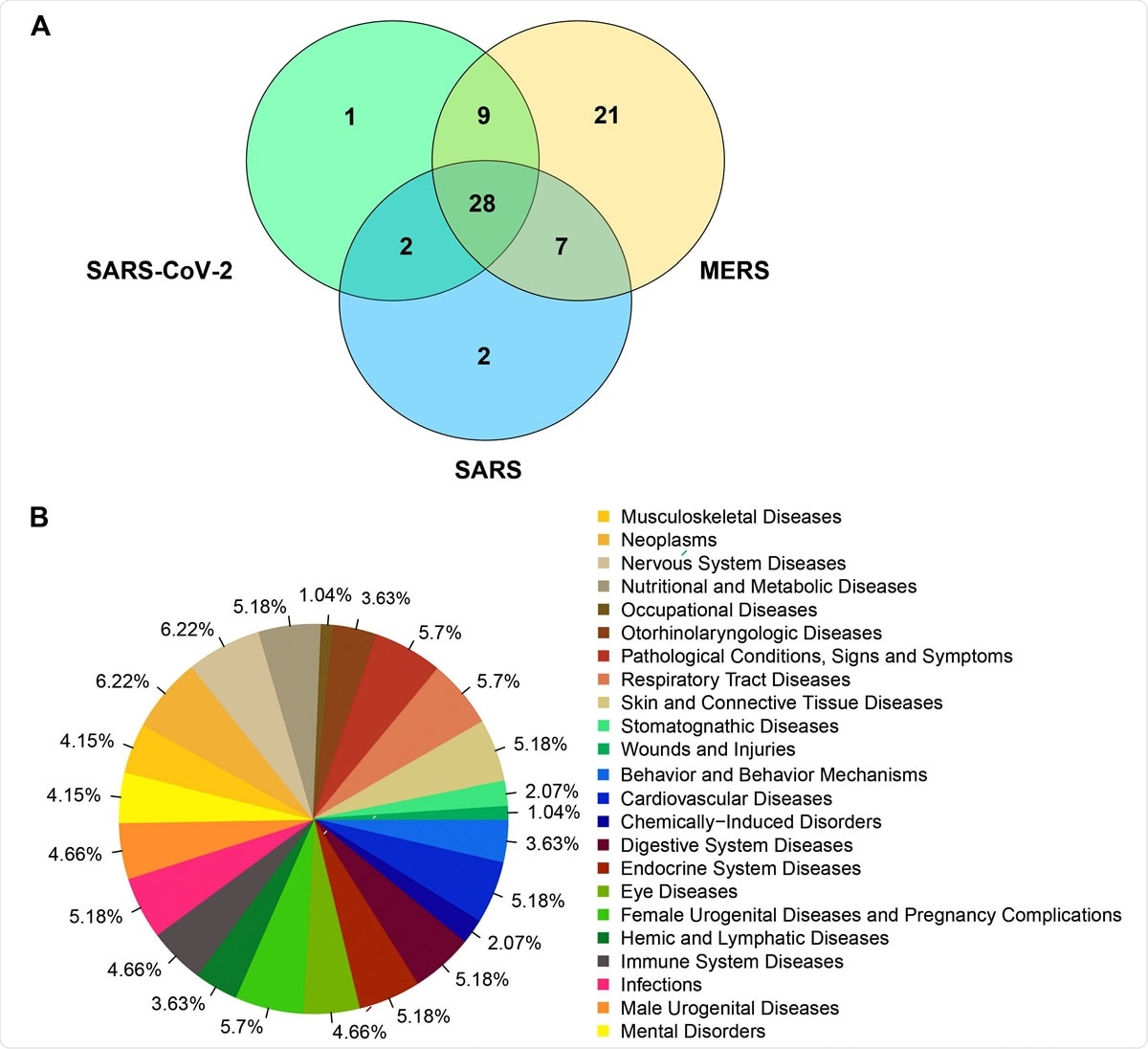
Venn diagram representing the number of human genes that clustered together with viral genes for SARS-CoV-2 (NC_045512), SARS (NC_004718) and MERS (NC_038294) based on the molecular features.B). Diseases frequencies associated to human genes grouped with viral genes of SARS-CoV-2, SARS and MERS in the clustering analysis.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Study: Identifying SARS-CoV-2-Human Molecular Matching
The current study by two researchers at the University of Buenos Aires aims to identify those viral molecular patterns and preferred codons, which reflect the host cell machinery and are, therefore, the preferred viral gene structure for optimal virus viability and human susceptibility. Scientists have suggested that codon pair bias and dinucleotide preferences are the primary factors that reflect the host’s codon usage, as has been proved with virus attenuation studies by codon pair deoptimization.
The scientists had the twin objectives of uncovering the molecular and adaptive nature of the three major human CoVs as well as to find out the host cell factors that selected for viral codons. Secondly, they tried to identify those viral genes that are essential for replication and the human genes required for the same process. This will help decide if population variability in genetic content models the gene features and helps develop host susceptibility.
The researchers looked at about 500 genome sequences downloaded from NCBI, including SARS, MERS, and SARS-CoV-2, classified by the host. Using only reference genomes, the quality was assessed, and the representative viruses were selected for further analysis.
The researchers then selected 463 highly expressed genes in the lung tissue of human hosts, with at least one-fold difference between their expression level here and in the tissue with the next highly-expressed level.
Codon Usage Bias Analysis
They performed codon usage bias (CUB) analysis using the total GC content of the CDS as well as that of the first, second, and third codon positions, denoted by P1, P2, and P3 respectively. They also calculated the codon indices such as relative synonymous codon usage (RSCU), the effective number of codons (ENc), codon adaptation index (CAI), codon bias index (CBI), the optimal frequency of codons (Fop), General Average Hydropathicity (GRAVY), aromaticity (Aromo), and GC-content at the first, second and third codon positions (GC1, GC2, and GC3), frequency of either a G or C at the third codon position of synonymous codons (GC3s), the average of GC1 and GC2 (GC12) and Translational selection (TrS2).
Using these, they were able to evaluate the degree to which specific codons were present (codon bias) for individual genes and for highly expressed genes, the frequency with which particular codons were expressed in a gene, the codon bias for different species, and the efficiency of codon-anticodon interaction. This allowed the determination of codon pair score (CPS) in coding sequences – “the natural logarithm of the ratio of the observed over the expected number of occurrences of a particular codon pair in all protein-coding sequences of a species.”
Meanwhile, the codon pair bias was used to find the CPS among the virus and host genes. In other words, the number of times a codon pair is expected to occur is a measure of the number of times it would occur without any association between the codons in the pair. A positive and negative CPS value shows that a particular codon pair is over- and underrepresented in the sequence of interest.
Thus, the CPS was calculated for each of the over 3,720 possible pairs of codons (61 x 61 codons). The Enc values were plotted against the GC3 values to show how G + C mutations affected the relationship between them, in contrast to the effect of selection pressure. Clustering methods were used to identify the groups of genes from the similarities in codon usage among human and virus genes.
The principal component analysis (PCA) was performed to find the most prominent factors that cause variation among the genes. Finally, they carried out a phylogenetic analysis on the DNA genome sequences of all the viruses to draw a phylogenetic tree.
Based on the fact that the human CoVs have CUB closely fitting the highly expressed proteins in the infected host tissue, the researchers examined gene molecules from SARS-CoV-2 and MERS as well as human genes. They found that in total, the mean Enc was similar among all the genes, viral and human, with only one unit of difference between the original non-human host and the virus.
Human SARS-CoV-2 Matches The analysis also showed that the human SARS-CoV-2 was different from that of bats and pangolins in the distribution of certain specific genes, depending mostly on the A/T content in P3.
The distribution of viral genes that are important to viral fitness, such as the M protein and the E protein, shared the tendency towards an A/T bias and showed a different distribution from that of non-human viruses. The CUB was higher for these genes compared to human MERS and SARS, though this contradicts the trade-off theory. An alternative explanation would be the effect of selection pressure that favors virus replication in a novel host, or by the recent jumping of the virus across species boundaries.
Implications of Matching Human-Viral Genes
The study thus suggests that virus replication in humans is easier with the clustering of the E protein with human genes that show molecular matching, which makes virion assembly and immunomodulation easier. This is supported by the positive CPB and a higher CPS correlation for the E protein-human gene clustering.
Similar patterns are seen with other genes like ORF6 and ORF8. A high CPB is seen with the N protein, ORF1ab, and the S protein. Changes in the GC3 position lead to synonymous substitutions and, in turn, optimization of codons in human cells, using the host cell machinery to translate only those genes that match the viral requirement at the molecular level.
Moreover, this could lead to a downregulation of human genes in the lung tissue, as has been reported to result from the imbalanced or wrongly modified tRNA expression. This, in turn, causes uncontrolled or abnormal protein synthesis, producing disease. This could also explain some effects of viral infection that are not because of direct viral injury.
The study concludes: “In our studies, we provided a list of human genes that could be particularly affected as a consequence of their molecular similarities with viral genes, not only belonging to SARS-CoV-2 but also SARS and MERS. The malfunction of these genes has been associated with different human pathologies and is in continuous increase.”
This could help to develop new preventives as well as to understand how human genes affect the probability of and the effects of COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources