The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic currently affecting the entire world is from a family of viruses that is common and found to infect only mammals and birds. There are 46 known species, with only 7 of them being capable of human infection. These include the SARS and MERS viruses, as well as the currently circulating SARS-CoV-2 virus. What makes some viruses able to infect humans while others remain confined to animals?
High infectivity of COVID-19
In comparison with either SARS or MERS, SARS-CoV-2 is a far more infectious virus in humans. The current study aims to understand where it came from and how it adapted to human hosts efficiently. This could help control the infection as well as evolve effective antiviral strategies and vaccines.
The virus that causes COVID-19 disease is very similar to the earlier virus that caused SARS, but even more identical to the bat coronavirus (CoV) named RaTG13 in the spike (S) gene sequence. The similarity is especially notable in the inter-gene regions. Still, there are some essential differences in vital genomic structures, such as the insertion of a polybasic furin cleavage site at the junction between the S1 and S2 subunits of the S protein.
.jpg)
Intermediate Horseshoe Bat (Rhinolophus affinis). Image Credit: Binturong-tonoscarpe / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Thus, even though most researchers support the identity of the bat as the reservoir for multiple CoVs, including SARS-CoV, the reservoir host of the current virus is unknown. The spike protein is key to the entry of SARS-CoV-2. It forms homotrimeric structures that protrude from the viral surface membrane. These are the recognition molecules for the host cell human ACE2 receptor.
Spike Protein and ACE2
This spike-ACE2 binding triggers membrane fusion and viral entry into the host cell. The spike protein is composed of two subunits, S1 and S2, with the receptor-binding domain (RBD) being located on S1. This binds to the ACE2 molecule. This activates conformational changes that lead to endocytosis of the virus and membrane fusion, releasing the virus to infect the host cell.
This key RBD sequence is the most diverse part of the viral genome and contains 6 amino acids, which are vital to binding the ACE2 receptor, and perhaps in crossing the species barriers to infect other hosts.
As with the earlier SARS-CoV, mutations have occurred in the RBD during its adaptation to different host cells along the transmission chain. The current study explores these mutations to understand this mechanism at the molecular level. In short, the binding capacity of the spike protein to the ACE2 receptor determines how easily the virus spreads across various species.
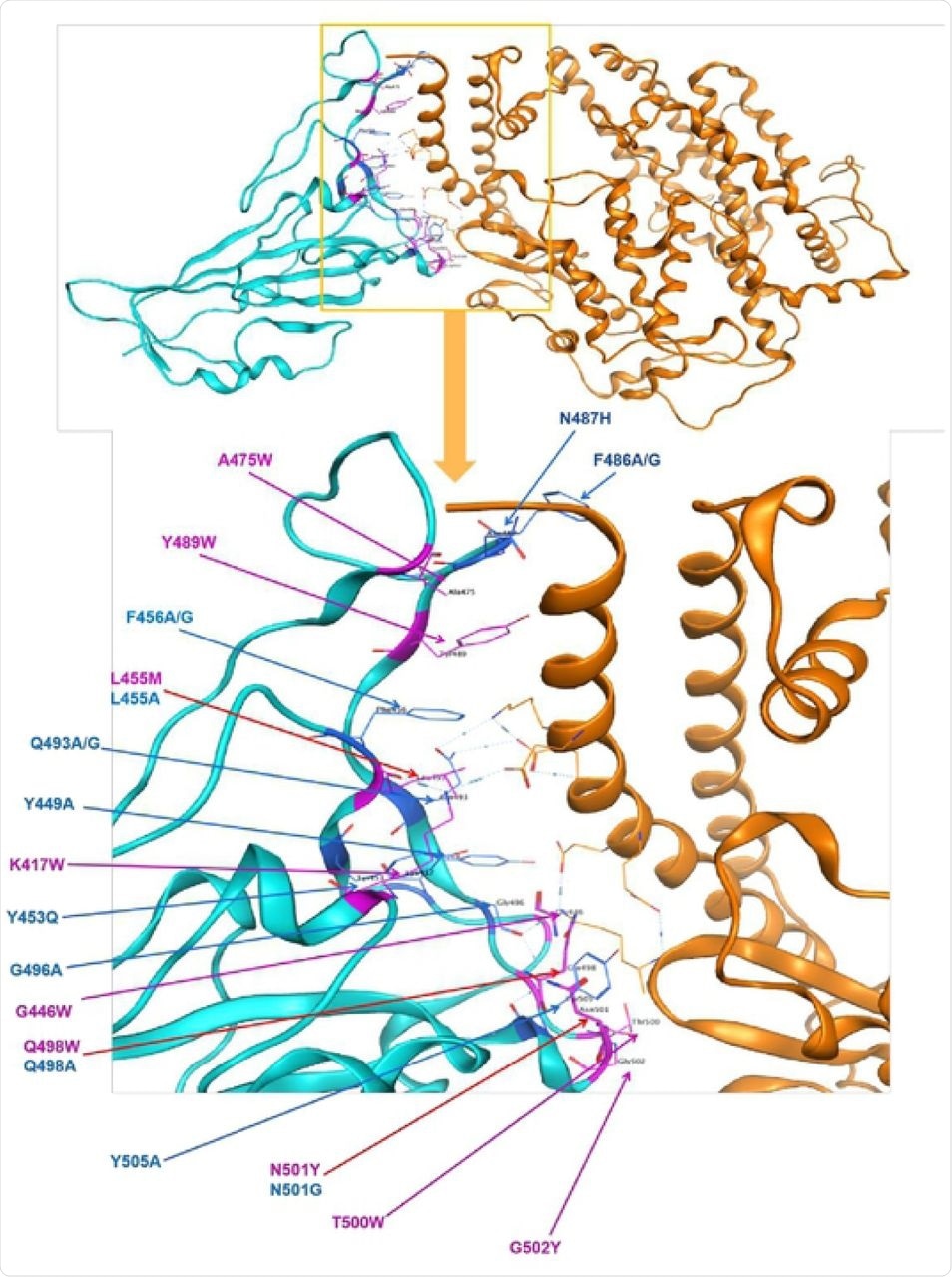
Identified polymorphism in spike protein RBD mapped to the structure of spike protein in SARS-CoV-2 in complex with ACE2 in humans. Cyan = spike protein, Orange=ACE2 direct bounding to spike protein. 23 point mutations causing affinity significant change on direct bounding of spike protein of SARS-CoV-2, blue presents decreasing affinity while purple shows increasing.
The Study: Tracing Adaptation via Structural Protein Mutations
The aims of the study are: the description of the phylogenetic relationship of different variants of SARS-CoV-2 in various populations, and the nucleotide polymorphisms from different regions of the viral genome, namely, the S, M, N, and E regions; develop a predictive method to find the S gene mutations that confer adaptive capacity on the virus, as well as the differences in mutation stability and binding affinity; and to determine the possibility of ACE2 variants that impair viral binding and hence its infectivity.
The study population comprised of 1,000 Chinese locals and other subjects. The ACE2 variants were analyzed to assess their binding capacity. The researchers also predicted the binding affinities of these variants to understand if those changes make them resistant or susceptible to the virus.
The study authors constructed a Maximum Likelihood (ML) phylogenic trees were constructed based on 2,147 genomes, which showed that it was closely related to the RaTG13 bat CoV and a pangolin CoV. Strikingly, at the molecular level, there is no close relationship between SARS-CoV-2 and SARS-CoV.
The researchers also reconstructed the tree based on the structural protein-encoding genes. They came to the conclusion that the evolutionary process might not be as important in deciding the phylogeny of these proteins compared to the adaptation of functional proteins.
S Protein Mutations and Their Effects
They found that there were 23 point mutations in the S gene that influenced the affinity and the stability of the S protein, 9 enhancing and 14 adversely affecting these properties.
They also looked at 1,150 variants in the S protein reported by the CDC and found that among 76 missense mutations in a single region, five enhanced its affinity binding, and only three increased the stability of the complex. Looking at the genome sequences that harbored these mutations, these were found to be close to the position of the SARS-bat and SARS-pangolin in the phylogenetic tree.
Earlier studies show that SARS-CoV-2 have higher affinity and viability than SARS-CoV in air and on surfaces, perhaps because the former could adapt to human spread better. The current findings confirm this with respect to the S protein.
Again, the finding that the SARS-CoV-2 S protein is most closely related to that of the horseshoe bat virus RaTG13, and then to the pangolin virus, agrees with earlier research, reinforcing the conclusion that the latter is unlikely to be the direct or only intermediate host of the virus. In fact, the insertion of the unique furin polybasic cleavage site insertion at the S1/S2 interface could have been acquired during passage through an unknown intermediate host.
The different variants of the spike protein observed in different countries suggest, on phylogenetic analysis, that the original infection could have occurred at multiple sites rather than only at Wuhan. The researchers caution that much more data is required to complete this direction of study, however.
Implications and Applications
They also found ACE2 variants, which, however, did not affect either affinity or stability. As a result, they suggest that “the mortality rate is also more likely to be related to medical and health conditions and the physical condition of patients” than to the ACE2 variant.
They conclude: “The analysis shared in this study would provide useful genetic information to track the mutations that occur in the spike protein of SARS-CoV-2 and prevent the recurrence of this epidemic, moreover, protect human beings from zoonotic coronavirus infection.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources