The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus that causes COVID-19 is a betacoronavirus, with a single-stranded RNA genome, and several structural proteins. Among these, the spike or S protein that binds to the host cell receptor, ACE2, is important as the antigen against which neutralizing antibodies are directed. A new study by researchers from the UK and Germany and published on the preprint server bioRxiv* in June 2020 reports the results of a detailed study of the S protein on the intact virus, using cryoelectron microscopy (cryoEM) and tomography. This could help understand the conformation of the S protein on the virion and how it interacts with neutralizing antibodies.
On cryoEM, the virus is a spherical particle, about 100 nm in diameter, with a dense viroplasm and with a lipid bilayer on both sides, on which there are spikes composed of the glycoprotein called the S protein. These recognize and bind to the ACE2 receptor on the cell surface, and are responsible for the endocytosis and subsequent viral entry into the host cell. This process involves a dramatic structural rearrangement, with quite distinct prefusion and post-fusion conformational states.
The infection of the virus in a human host cell leads to a profound remodeling of the organization of the internal membranes of the host cell. This leads to the production of organelles that allow viral replication. The viral structural proteins are incorporated into the endoplasmic reticulum (ER), from where they are transported to the ER Golgi intermediate compartment (ERGIC). The genome is then wrapped around by the N protein capsid, which results in the formation of viral particles that are then transported to the surface of the cell and released.
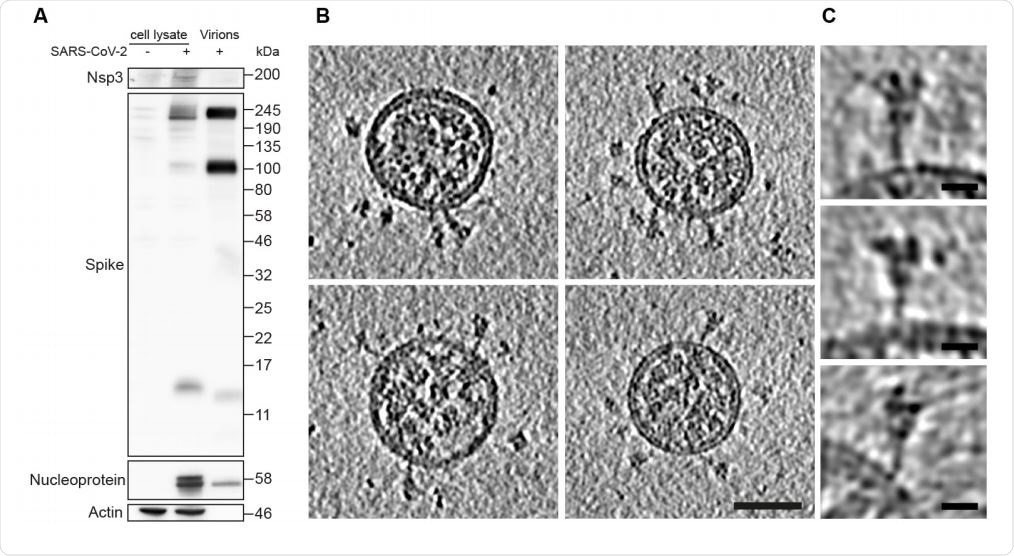
Characterization of virus production and representative images of intact, authentic SARSCoV-2 virions. (A) Western blot analysis of SARS-CoV-2 S and N in lysates of VeroE6 cells and in virus preparations. In released virions, S is present in both cleaved and uncleaved forms. (B) Four representative tomographic slices of SARS-CoV-2 virions from the supernatant of infected cells. Virions are approximately spherical, contain granular density corresponding to N-packaged genome, and have S protein trimers protruding at variable angles from their surfaces. Scale bar 50 nm. (C) Three example S trimers from the dataset shown as projections through the trimer to illustrate the variable tilt towards the membrane. Scale bar 10 nm.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Prefusion Spike Structure
The prefusion structure of the S protein is usually studied using secreted S protein expressed on other cells, which is then purified and then subjected to cryoEM and reconstruction of single particles. This has led to the following understanding: the prefusion S protein has a receptor-binding domain (RBD) atop a broad spike composed of three spike protein units, above the fusion core. The three copies of the RBD, one on each spike protein, sit amidst the three N-terminal domains (NTDs).
When the prefusion conformation is in the closed state, all the three copies of the RBD are flat on the surface of the spike, blocking the receptor binding site. In the open state, one or more of the three RBDs are exposed to allow the receptor to bind to the ACE2.
The spike trimer is glycosylated, and there are 22 N-glycosylation sites on each protomer. When the prefusion state gives way to the post-fusion state, the fusion peptide and the transmembrane domain come together at one end of the long needle-like structure, along which there are five N-linked glycans.
The Study: Spikes in situ on the Cell Surface
The current study looked at SARS-CoV-2 viral particles retrieved from the infected cells, without either the concentration or the purification step. The researchers saw that the particles were similar in shape and size to the earlier description. The protruding S trimers had two forms. One was the thin structure resembling the needle-like post-fusion structure, while most were wider, resembling the prefusion form. About 97% were prefusion and 3% post-fusion.
This agrees with tomograms of the virions produced by infected cells in culture, which also show the same type of spike proteins, about 25 per virion. The researchers also examined the thin and wide S trimer shapes and found that they showed a striking correspondence to the post-fusion and prefusion states, respectively.
RBD and S Trimer Classification
The RBDs in each of the protomers were classified individually. There were three kinds of classes, closed RBD, open RBD, and mostly closed but with somewhat weakened density, which could indicate that the conformation could be more variable. They found that closed trimers and trimers with only one RBD open had the same frequency, about 40% to 45%. About 13% had two RBDs open. These findings show that the RBD opening is also seen on the viral surface as well as in recombinant S trimers.
Secondly, they classified the trimers by orientation and found that the stalk region attaching the spike to the membrane acts as a hinge, allowing the spike to tilt in all directions. They also found that the S protein occurs at a distribution of about 1 per 100 nm2, which suggests that avidity is less important in receptor binding than for influenza viruses.
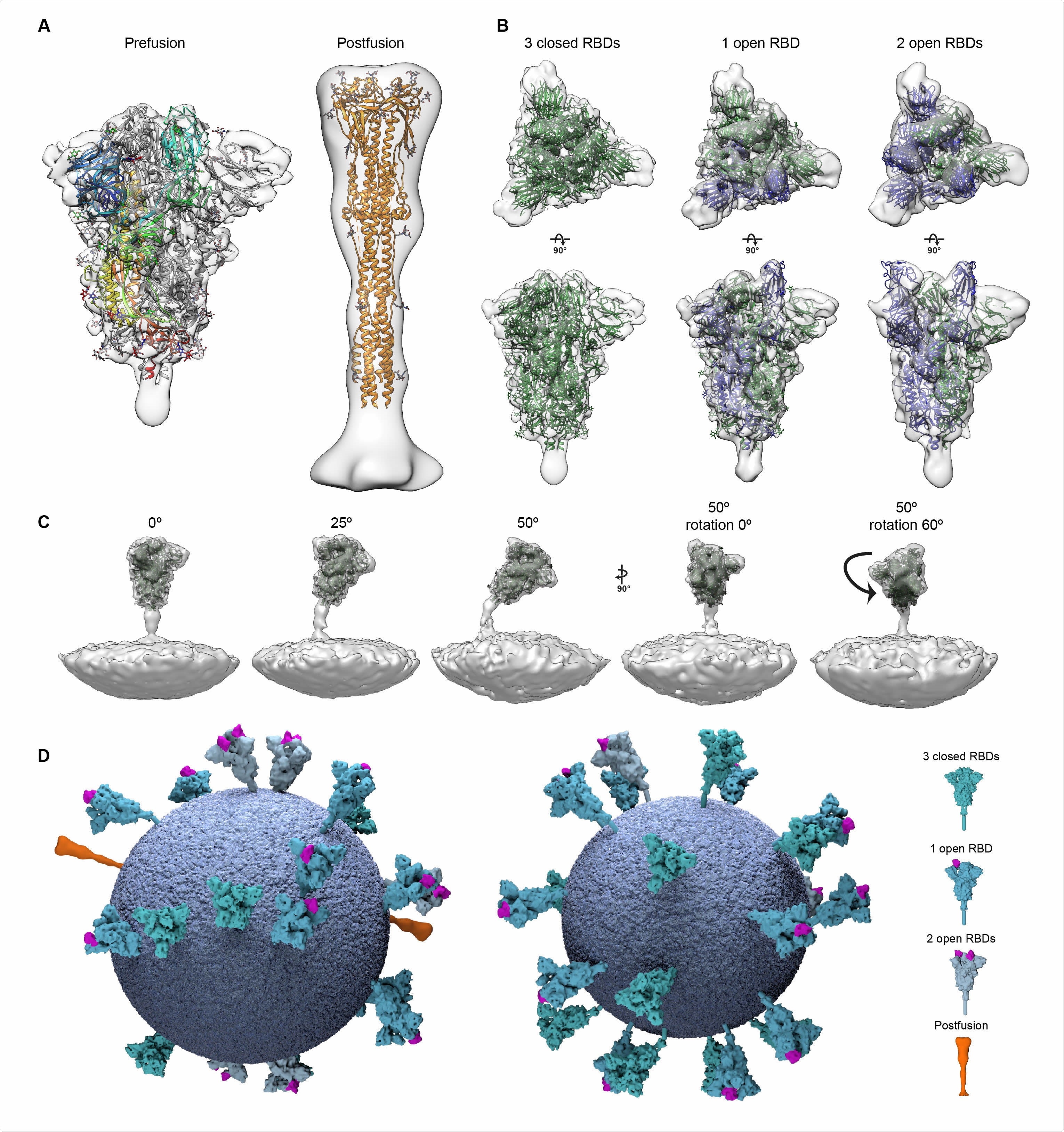
Structural analysis of SARS-CoV-2 S trimers from intact virions. (A) Structures of the prefusion (left) and postfusion (right) S trimer from intact virions determined by subtomogram averaging. The structures are shown as a transparent grey isosurface fitted with structures of the closed, prefusion SARS-CoV-2 S trimer (PDB 6VXX) and the postfusion SARS-CoV-1 S trimer (PDB 6M3W). In the prefusion form, one monomer is colored from blue (N terminus) to red (C terminus). The N-terminal domain is blue, the RBD appears cyan. Note that the NTD does not fully occupy the EM density because a number of loops are not resolved or built in PDB 6VXX. (B) The different conformations of the prefusion S trimer observed on intact virions by subtomogram averaging. Three conformations were observed: all RBDs in the closed position (left, fitted with PDB 6VXX); one RBD in the open position (center, fitted with PDB 6VYB); two RBDs in the open position (right, fitted with PDB 6X2B which does not include modeled glycans). Note that the twoopen conformation has only been observed in vitro after inserting multiple stabilizing mutations. S monomers with closed RBDs are green, and with open RBDs are blue. (C) Averaging of subsets of trimers grouped according to their orientation relative to the membrane shows flexibility in the stalk region. Examples are shown for pools centered at 0°, 25° and 50° from the perpendicular, and for two different rotations of the trimer relative to the tilt direction. (D) 3D models of two individual SARS-CoV-2 virions with a membrane (blue) of the measured radius, and all spike proteins shown in the conformations, positions and orientations determined by subtomogram averaging.
Implications of the Study
Notably, the researchers found that following concentration, the cryoEM images showed S trimers in only the closed conformation, both pre- and post-fusion, unlike other studies that suggest that the open RBD in the soluble S protein trimer is found in many different positions.
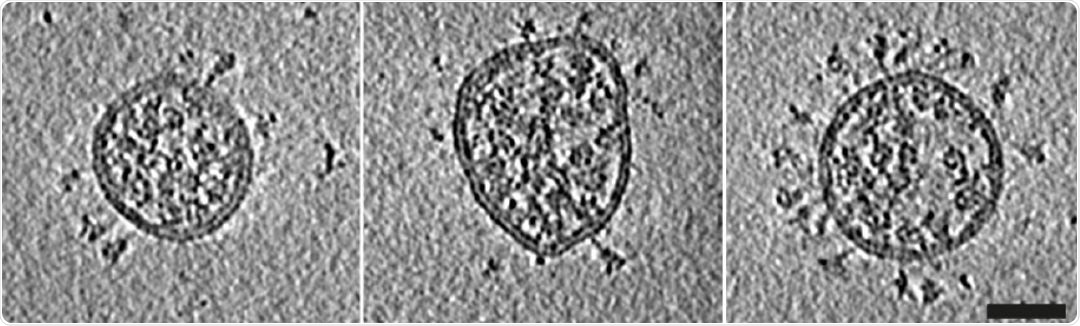
Central slices through representative viruses. Virions from Calu-3 cells had a slightly broader diameter distribution than those from VeroE6 cells. Scale bar 50 nm.
This leads the researchers to suggest, “The open prefusion conformations of the spike protein we observe in situ, but not after concentration, are fragile and may be affected by purification procedures, which has implications for vaccination strategies.”
The structure of the S trimer in situ, on the surface of the virion, was derived, showing the position of 17 of the 22 predicted N-glycan sites on the monomer, with the other 5 being in the stalk or on the NTD.
They comment, “Overall, our structure is very similar to that previously published for the soluble trimeric ectodomain in the closed prefusion form stabilized by a double proline mutation. This provides an important validation of the use of recombinant, purified S protein trimers for research, diagnostics, and vaccination.”
The study shows that cryoEM can be used to study antibody-Spike binding on the viral surface, a vital step in discovering how neutralizing antibodies, and those to the S2 subunit and the stalk region, in particular, inhibit viral infection, and in shaping preventive vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ke, Z. et al. (2020). Structures, Conformations and Distributions Of SARS-Cov-2 Spike Protein Trimers on Intact Virions. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.27.174979. https://www.biorxiv.org/content/10.1101/2020.06.27.174979v1
- Peer reviewed and published scientific report.
Ke, Zunlong, Joaquin Oton, Kun Qu, Mirko Cortese, Vojtech Zila, Lesley McKeane, Takanori Nakane, et al. 2020. “Structures and Distributions of SARS-CoV-2 Spike Proteins on Intact Virions.” Nature 588 (August): 1–5. https://doi.org/10.1038/s41586-020-2665-2. https://www.nature.com/articles/s41586-020-2665-2.