The current COVID-19 pandemic is caused by a novel coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is part of a family of enveloped viruses with a large single-stranded RNA genome. The nucleic acid is compactly packed in an indistinct structure called the nucleocapsid, made of N protein. Now, a new study by researchers at the University of California San Francisco and published on the preprint server bioRxiv* in June 2020 shows the effect of phosphorylation on the state and function of the N protein.
When SARS-CoV-2 enters a host cell, it is disassembled, the nucleocapsid breaks apart and allows the genome to be translated into several non-structural proteins, including RNA-dependent RNA polymerase. These proteins rearrange the intricate membranes of the endoplasmic reticulum of the host cell to form the replication transcription complex (RTC). The RTC is a framework on which the viral replication and transcription proteins are arranged, and perhaps hidden from the host’s innate immune response.
The replication of the viral genome begins when the RNA strand is used as a template to produce a complementary strand in the subgenomic regions that encode four major structural proteins, S, M, N, and E, required for viral assembly. The mechanism of subgenomic transcription involves the completion of genes encoding structural proteins one by one, with each gene transcription ending as the polymerase skips intervening regions to a transcription-regulating sequence (TRS) at the 5’ end. This results in the formation of subgenomic fragments.
These fragments are subjected to transcription, to produce the original viral genes. These can be translated to produce the structural proteins needed to reassemble the virus.
N Protein Phosphorylation Necessary for Viral Transcription
The most abundant subgenomic proteins encode the N protein, and its translation proceeds at high levels early in the course of the infection. It is therefore found to pile up in clusters at the RTC and may be instrumental in bringing about the structural rearrangements in the RNA that is are needed for subgenomic transcription to occur.
The N protein is found in almost the same form among all coronaviruses and has two globular domains, the N-terminal and C-terminal domains (the NTD and CTD, respectively). Around these are intrinsically disordered regions. The N protein is dimeric, and the structure has multiple sites to which RNA binds, including a sizeable RNA-binding groove formed by two tightly bound CTD, and another on the NTD. In some conditions, these structures form oligomers by binding between different regions of the protein.
In the disordered region, there is a sequence with abundant serine-arginine (SR) that is thought to be essential for the regulation of N protein function. Early in the course of the infection, this undergoes rapid phosphorylation at many sites, mediated by cytoplasmic kinases. This facilitates subgenomic transcription in the RTC. As infection proceeds, nucleocapsid synthesis and viral assembly become independent of N protein phosphorylation.
The current study is aimed at understanding how the N protein is influenced by phosphorylation.
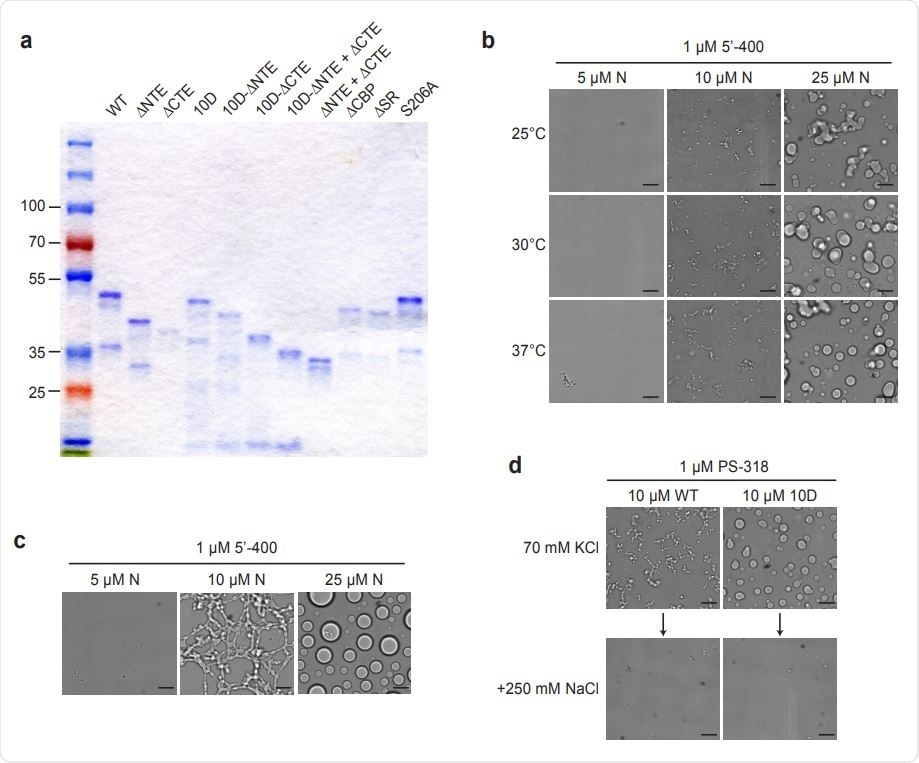
Characterization of N protein condensates. an SDS-PAGE analysis of all N protein mutants used in this study, stained with Coomassie Blue. b, N protein was incubated at the indicated temperature for 30 min in the presence of 1 µM 5’-400 RNA. Scale bar, 10 µm. c, N protein was incubated with 1 µM 5’-400 for 16 h at room temperature. Scale bar, 10 µm. d, Condensates of 10 µM WT or 10D N protein were formed in droplet buffer (70 mM KCl) by incubation with 1 µM PS-318 RNA for 30 min and imaged. NaCl was then added to a final concentration of 250 mM for 15 min before imaging again.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Oligomerization of N Protein Requires RNA
Under some conditions, the N protein forms oligomers. This appears to be liquid-like droplets, but in order to remove the possibility of RNA contamination, the researchers removed the RNA. They then found few microscopic protein structures but enhanced protein structure formation once RNA was added. The new structures closely resembled the native forms. In other words, RNA is needed for higher-order oligomers to be formed.
With the addition of RNA at room temperature and at a concentration of 10 μM, the N protein formed networks of small “liquid-like beads” in the presence of subgenomic RNA. As the N protein concentration rose, the droplet diameter increased to several microns. The control RNA, however, caused the formation of clusters of amorphous strands, which were only partially liquid-like in appearance.
When RNA concentration was low, small spherical structures were formed, but at higher RNA concentrations, filaments were formed. When RNA and N protein concentrations were almost equal, no structures were generated. The researchers conclude, “These results suggest that condensates depend on the crosslinking of multiple N proteins by a single RNA.”
Next, they found that when they added a TRS segment of RNA 10 nucleotides long, droplets were formed rapidly. With longer RNA segments, filaments were formed instead, to generate a gel-like structure.
At different N protein-TRS concentrations, the formation of droplets showed a marked change, from a few droplets at high N concentration to abundant formation at low N protein levels.
This indicates that in the absence of long RNA, the N protein inhibits the formation of TRS-bound oligomers. In other words, monovalent TRS-N protein binding produces a change in the N protein structure. The increased number of low-affinity protein-protein interactions causes droplets to form. However, when long RNAs are present, as, under physiological conditions, multivalent RNA-N protein binding enhances droplet formation.
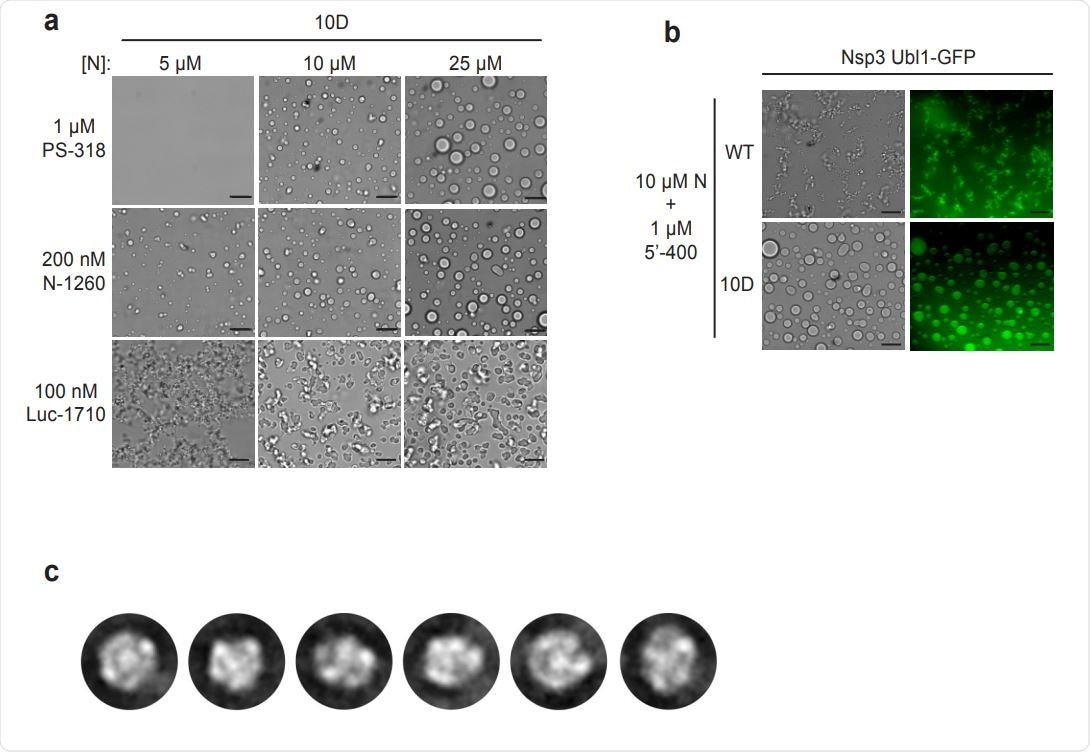
Characterization of N protein condensates. a, Images of N protein 10D mutant following 30 min incubation with the indicated RNAs. Scale bar, 10 µm. b, 10 µM wild-type (WT) or 10D N protein was incubated with 1 µM 5’-400 RNA for 10 min. Nsp3 Ubl1- GFP was then added to a concentration of 1 µM and incubated for an additional 15 min before imaging in brightfield (left) or fluorescence (right). c, 2D class averages of particles from the EM analysis of wild-type N protein and PS-318 RNA shown in Fig. 4c.
Phosphorylation Changes N Protein Condensate Form
Similar experimental variations showed that the N protein and the viral RNA work together to assemble into a variety of structures depending on the very different intramolecular and intermolecular forces at work.
The unmodified N protein is a gel-like filamentous condensate, showing a partially ordered structure. This is because filaments are more rigid and probably form after high-avidity interactions, such as multivalent RNA-protein and protein-protein binding. The presence of long RNA sequences increases the protein-protein interactions because they can bind to the many RNA binding sites on the protein.
The findings also seem to indicate that when the SR region of the genome is phosphorylated, the condensates change to become more liquid. Both protein-RNA and protein-protein binding via this region are blocked, as well as intramolecular binding with RNA at other sites. This resulting loss of affinity would give rise to a more liquid droplet.
Two Functions, Two Forms
Phosphorylation N protein oligomers thus exist in different forms, which are required for its two primary functions of compact RNA packaging and subgenomic transcription. With the first, the N protein forms a structural framework. This allows nucleocapsid assembly, which may rest on a foundation of uniform droplets of protein and RNA.
For the latter purpose, the liquid-like attributes of phosphorylated N protein droplets could be vital at the RTC. The N protein undergoes complete phosphorylation as soon as it is synthesized, and rapidly forms a structure in association with large membrane structures thought to form the RTC.
Phosphorylation of N protein is necessary for its subgenomic regulatory function at the RTC. Putting all this data together, the researchers say their findings suggest the formation of a protective compartment for viral replication and transcription components, formed by a liquid-like phosphorylated N protein matrix with RNA loosely bound to it, and linked to the membranes of the RTC by nsp3.
Inhibitors such as the small molecules that inhibit GSK-3 kinase, a key phosphorylation enzyme, prevent the normal processing of mouse hepatitis virus (MHV) genomes and lead to a fall in the production of virions in the cells infected by these viruses. They could perhaps play a role in preventing the progression of early COVID-19 infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources