The current pandemic of COVID-19 is caused by the severe acute respiratory syndrome coronavirus (SARS-CoV) -2, leading to severe pneumonia, acute respiratory distress syndrome, and death in a sizable minority of cases. However, the most significant number are asymptomatic or have only mild infection from which they rapidly recover.
Different Manifestations, Different Immunity
The range of manifestations of COVID-19 includes asymptomatic infection, mild fever, and cough, mild pneumonia, severe pneumonia, and Acute Respiratory Distress Syndrome (ARDS). The various factors involved in this diverse range of symptomatology include impaired immunoregulation, the cytokine storm, and the decreased number of immune cells.
T cells are adaptive immune cells that recognize antigenic peptides presented by specialized cells bearing HLA I or II molecules. The peptides presented on cells that have HLA I molecules on their surface are recognized by CD8 cytotoxic T cells. Still, those with HLA II molecules present antigens that are recognized by CD4 T helper cells.
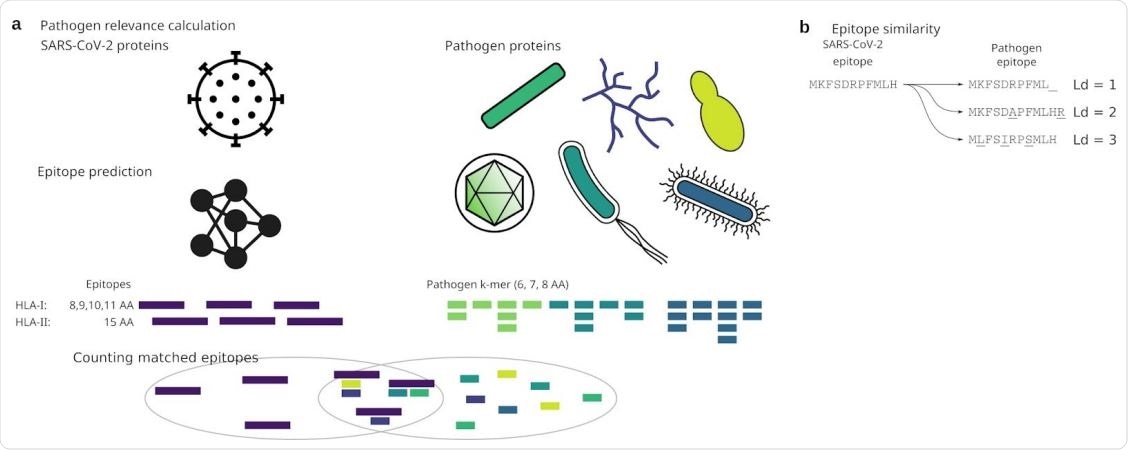
Analysis approach

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Cross-Reactive T Cells
The current study was motivated by the finding of specific T cells targeting the SARS-CoV-2. In other words, the immune response to the SARS-CoV-2 virus involves T cells, but healthy individuals are also able to respond to peptides derived from the viral S protein. This may mean that they already have preexisting immunity to this virus. The source of such exposure is unknown, though one epitope common to influenza and SARS-CoV-2 viruses has been found.
The researchers consider that the endemic human coronaviruses (CoVs) are likely to have induced this immunity to the current virus since their genomes have a high degree of similarity to it. However, earlier serologic studies have reported that the antibody response to these endemic viruses wanes after 4-12 months. It could also be possible that the same CoV can cause reinfection within this period. Overall, these endemic CoVs do not seem to generate lasting immunity.
The correlation between antibody and T cell response is not known. The genetic drift in endemic CoVs occurs at a slower pace than for influenza viruses A and B, which could indicate that the presence of T cells reactive to the S protein of the current SARS-CoV-2 was due to other common pathogens, and not just the endemic CoVs.
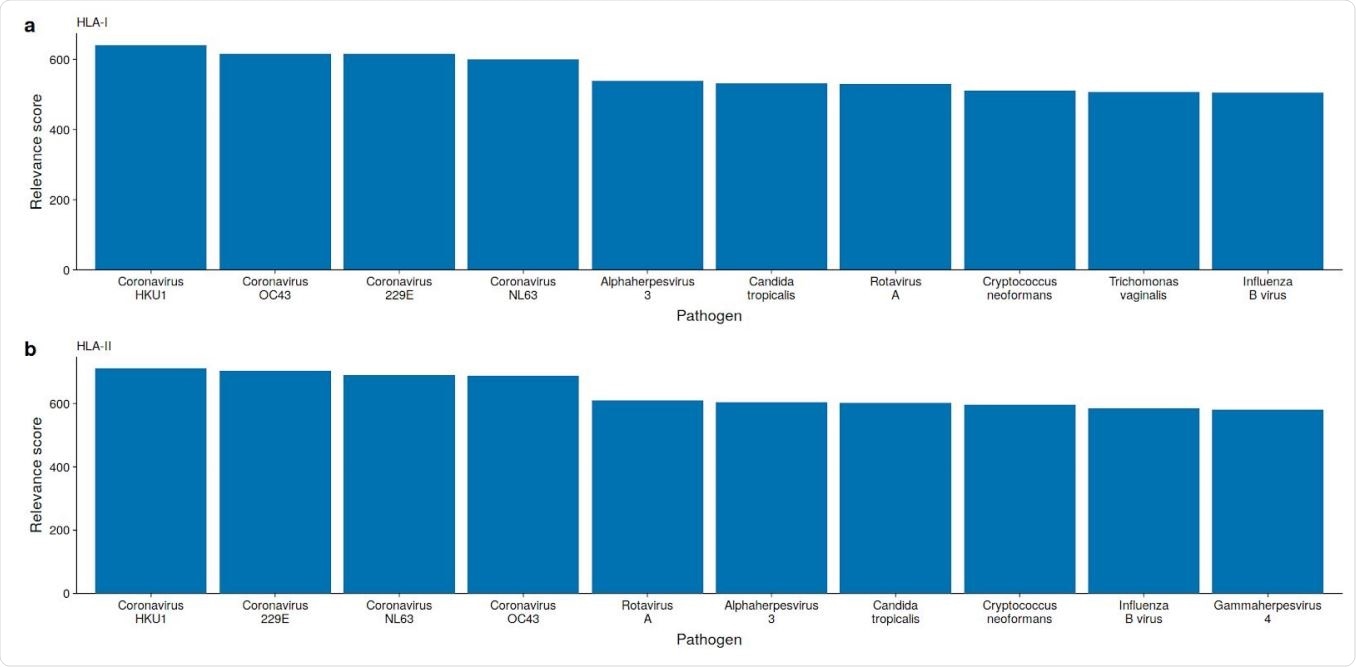
Some viruses and bacteria have peptides (k-mers) matching SARS-CoV-2 epitopes
The Study: Finding Epitopes Similar to SARS-CoV-2 Epitopes
The researchers carried out an in silico analysis of specific epitopes supposed to be important in the pathogenicity of the virus. Despite some dissimilarity, the varicella-zoster virus (VZV) has the highest number of epitopes predicted for the SARS-CoV-2 virus, but only those which bind to HLA class I molecules, especially HLA-B and HLA-C.
The researchers said, “We find the highest number of similar SARS-CoV-2 HLA-I epitopes in VZV, followed by OC43, and HKU1 with 1292, 1189, and 1163 epitopes, respectively.” HLA-II binding epitopes did not show this similarity.
However, the fact remains that T cells reactive to the SARS-CoV-2 comprise both the CD4 and CD8 cells, those which react to antigens bound by HLA-I or HLA-II classes. This means that the VZV is unlikely to be the only pathogen behind the cross-reactive T cells. Instead, the highest number of epitopes in common are found among the endemic CoVs, especially the beta-CoVs.
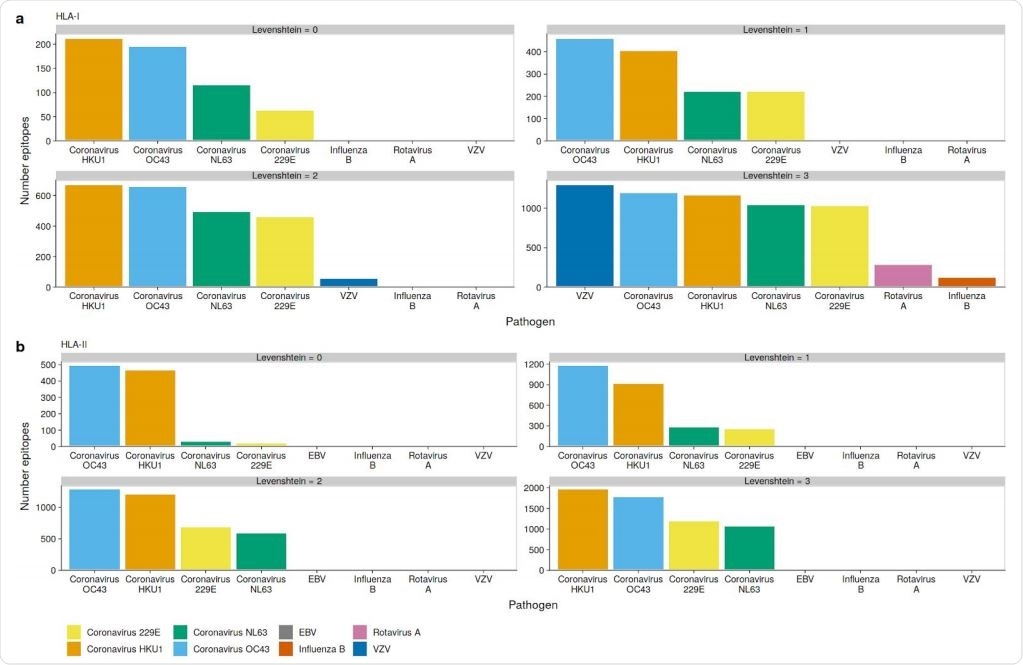
OC43 and HKU1 epitopes can be presented on many HLAs
Limitations and Conditions of the Study
The epitopes that bind to HLA-I molecules were predicted with high accuracy, but those which bind to HLA-II molecules not so much. This could be due to the fact that HLA-II is in the form of a dimer that forms the peptide-binding groove, leading to over 4,000 different possible combinations, compared to the far fewer combinations possible with the HLA-I and β2-microglobulin chains.
The current study focused on the HLA sets most common in Europe, avoiding those which include laboratory model antigens, such as the human autoantigen and those from the parasite Trypanosoma cruzi.
Using a smaller amino acid set for epitope identification helped compare sequences without the need for prior alignment since the k-mer nucleotide sequence is part of the target protein. While this might alter the results, it is unlikely since the anchor amino acids are deep within the HLA molecule.
Implications
The findings mean that the betacoronaviruses OC43 and HKU1 are most probably responsible for the preformed T cell immunity specific to SARS-CoV-2 displayed by the uninfected individuals in this study. This also means that these viruses are capable of eliciting durable or permanent T cell responses, given the lack of identity between the epitopes of these different viruses.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources