The most fatal form of COVID-19 disease is acute respiratory distress syndrome (ARDS), caused by direct viral injury as well as indirect noxious effects caused by cytotoxic chemicals and inflammatory processes. A new study published on the preprint server bioRxiv* in June 2020 shows that this is partly due to the rapid onset of inflammation caused by the infection of type 2 alveolar cells with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The study of the human lung tissue by in vitro models is essential to understand how respiratory pathogens target these cells, especially since it is now becoming clear that many different epithelial cells in the respiratory tract express the host cell receptor, angiotensin-converting enzyme 2 (ACE2), thus allowing infection with the virus, as is seen in living organisms as well.
The lung phenotype of COVID-19 is characterized by viral targeting of the lung epithelium, especially the type 2 alveolar epithelium (AT2s). Not much is known about how the lung alveoli respond to the infection since these cells are difficult to obtain for study.
The Study: In Vitro Model of Early COVID-19
The current study reports the findings of an in vitro human model of the early stage of SARS-CoV-2 infection of the alveolar lung epithelium, using AT2s derived from induced pluripotent stem cells – iAT2s. The cell cultures were engineered to provide a faithful representation of the infected cell layer in the lung, as a cell monolayer, all pointing one way and with an intact barrier membrane, as well being specific for the genes found only in AT2s.
Rapid Profound Shift in Gene Expression and Transcription
The researchers found that at one day after infection with SARS-CoV-2, the virus causes a rapid shift in transcription in the infected iAT2s. More than 4,500 genes were expressed at different levels in the infected cells compared to the controls on the first day post-infection.
This difference was seen in over 10,700 genes when infected cells on day 1 were compared to those on day 4 post-infection, and a similar number of genes were differentially expressed when infected cells as a whole were compared to controls.
Viral transcripts made up the most significant number of differentially expressed genes at both time points, at up to a third of all reads on day 1 post-infection. Genes specific to AT2s were those most frequently lost, increasingly downregulated from day 1 to day 4.
Progressively Increasing Inflammation
Infected cells showed increasingly higher levels of inflammation at both points, and indeed over the whole course of the infection, mediated byTNF and NFkB.
In this picture, inflammation takes hold, characterized by cytokines released in response to the activation of genes by NF-kB. At four days from infection, epithelial interferon release occurs in a delayed manner, and rapid dedifferentiation of the mature lung alveolar epithelium, accompanying this change.
Increasing Cytotoxicity
With the progress of the infection, infected iAT2s show signs of toxic changes such as increased apoptosis and stress-linked signals. These were seen by the fourth day from infection, resulting in death. Since these are important for the renewal of the alveolar cells, this is a crucial change. Lung tissue histology also showed the same pattern of regional injury to the alveolar epithelium and hyaline membrane formation.
Drug tests on the recently developed camostat mesylate with the same type of cell showed that inhibition of the TMPRSS2 protease was an effective strategy, supporting the hypothesis that it played an essential role in viral entry into the host cell.
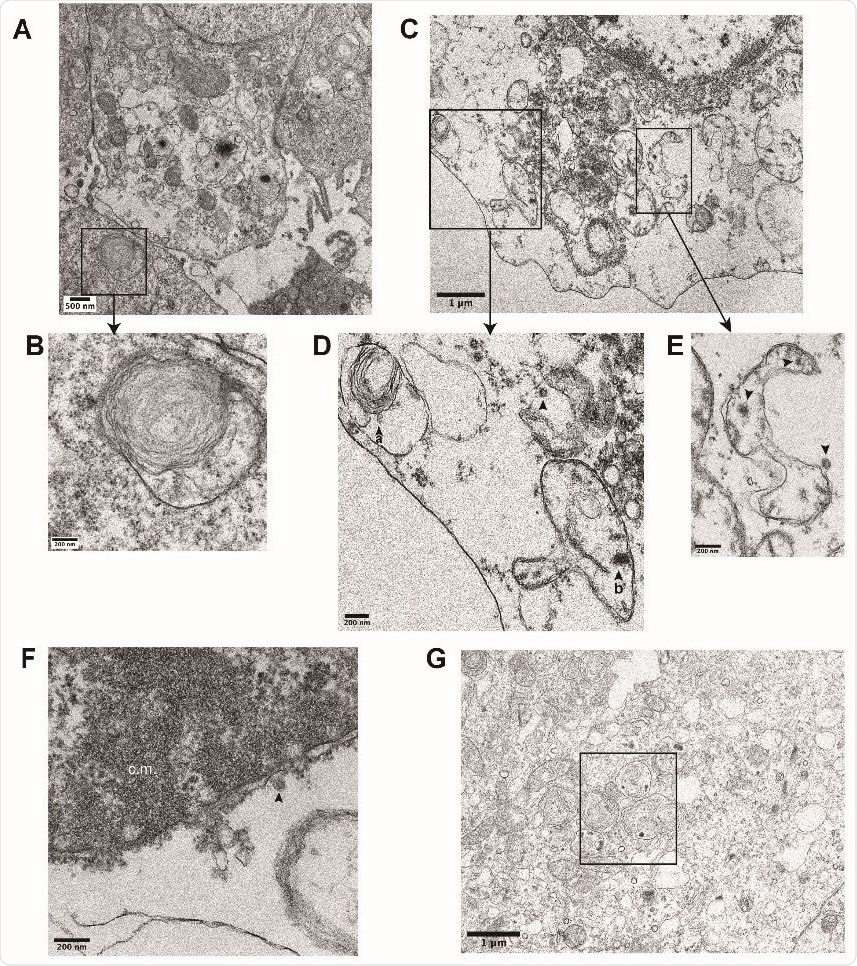
Ultrastructural analysis of iAT2s infected with SARS-CoV-2. (B) Transmission electron micrographs of mock-infected iAT2s at ALI (A-B) demonstrating lamellar body expression but no detectable virions. iAT2s at ALI infected with SARS-CoV-2 at an MOI of 140 and fixed 1 dpi (CG) contain visible virions (C-E, G, arrowheads) in the cytoplasm (D,E), within lamellar bodies (D, arrowhead a) (G, see Fig. 2J for inset), and within double-membrane bound structures (D, arrowhead b) (E, arrowheads). Virions are also found extracellularly (F, arrowhead) and some iAT2s contain convoluted membranes (F, c.m.).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Implications and Applications
The use of iAT2s in a form that can be used in 2D models of acute lung injury is an important adaptation, as it “allows straight-forward simulations of apical viral respiratory infections of a self-renewing cell type that can be scaled up, nearly inexhaustibly, and studied in highly pure form, thus simulating cell-autonomous or “epithelial only” host responses to pathogens.”
The ability to achieve adapted iAT2s with a higher than the typical expression of maturation genes shows that the study was carried out on cells, very like the real-life targets of infection. The observation of viral particles within lamellar bodies, which are the organelles that package surfactant, may indicate that this is a site-selective for the virus and perhaps one which is disrupted by it. This organelle is found only in mature AT2s, and not in lung cell lines typically used for cell culture experiments.
The findings show that AT2s respond to SARS-CoV-2 infection by inflammatory signaling within 24 hours, predominantly via the TNF-NFkB signaling pathways. This results in a loss of surfactant, cytotoxicity, and cell death that are probably a reflection of the real-life response of the lung to this infection. The same findings were obtained from lung autopsy.
IFN responses are not significant compared to the former pathways, but the virus does respond rapidly to treatment with IFNλ and IFNβ. This could allow AT2s to be treated at milder stages of the infection before they progress to ARDS. This finding is, therefore, clinically relevant. Moreover, the delayed IFN response is common to airway epithelium infected with SARS-CoV and MERS-CoV as well and is one of the risk factors for disease severity in SARS.
The researchers sum up: “Our model system reveals the cell-intrinsic responses of a key lung target cell to infection, providing a platform for further drug development and facilitating a deeper understanding of COVID-19 pathogenesis.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources