The COVID-19 pandemic is taking thousands of lives, and the worldwide incidence is measured in hundreds of thousands. Antivirals and vaccines are being urgently sought to counter this growing threat. Now, a new study of the structural dynamics of the SARS-CoV-2 spike protein published in the preprint server bioRxiv* in July 2020 reports on the epitopes that could be possible immune targets for the development of vaccines.
The abundant glycan residues of the spike protein prevent antibodies from targeting it and hinder the access of drugs as well. Therefore, the current study was aimed at capturing the conformational changes of the Spike (S) protein and the dynamic change in the glycan coating. The researchers used molecular dynamics (MD) simulations to paint a vivid picture of the glycosylated S protein and identify candidate immunogenic epitopes, especially in the S2 subunit, which is responsible for the fusion of the virus to the host cell.
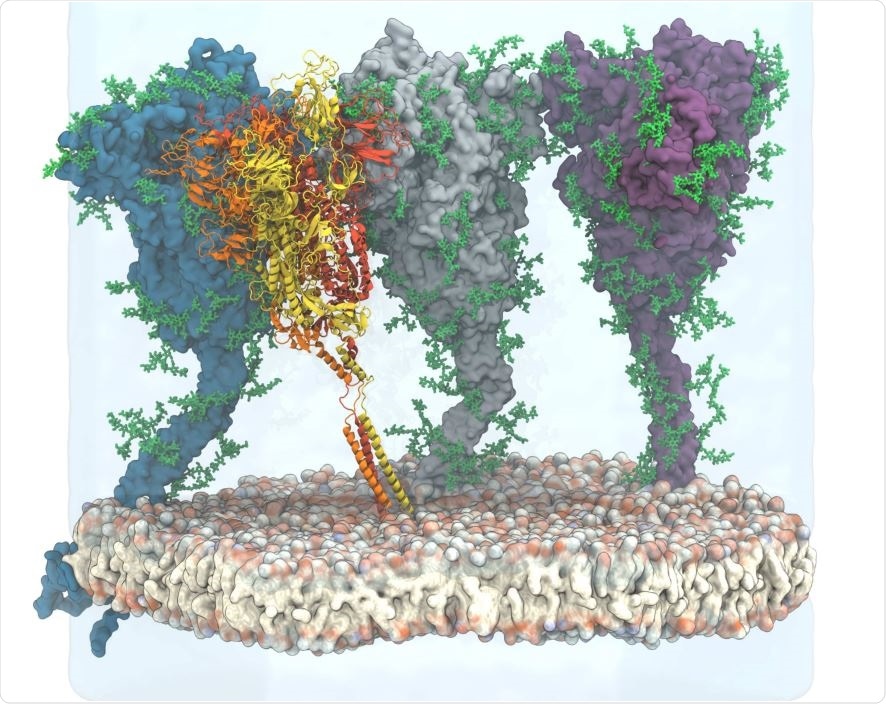
View of the simulated atomistic model containing four glycosylated and membrane-anchored S proteins in a hexagonal simulation box. Three proteins are shown in surface representation with glycans represented as green sticks. One protein is shown in cartoon representation, with the three chains colored individually and glycans omitted for clarity. Water (transparent) is only shown for the lower and back half of the hexagonal box, and ions are omitted for clarity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Spike Protein Accessibility
Antibodies need to have access to epitopes to bind to them. The current analysis showed that the dense shield of glycans significantly reduced the accessibility of S protein to antibodies. At any given time, only a small part of the surface of the spike is actually covered by these sugar residues, but they are so dynamic that they effectively shield the entire surface. This was seen by both ray analysis and Fab docking studies.
The most significant shielding effects are seen in the part nearest the membrane, where the non-glycated HR2 coil is open to the antibody attack almost entirely. Still, with glycosylation, it becomes inaccessible to the Fab fragment and other large molecules.
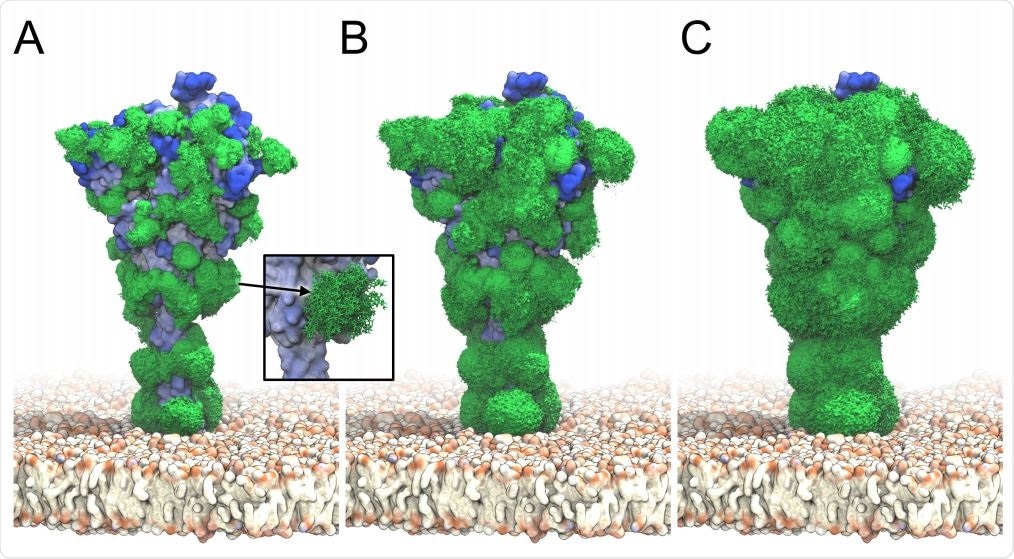
S glycan dynamics from ˜ 2 µs MD simulations. Timeaveraged glycan electron density isosurfaces are shown at high (A), medium (B), and low (C) contour levels, respectively. The blue-to-white protein surface indicates high-to-low accessibility in ray analysis. (Inset) Snapshots (sticks) of a biantennary, core-fucosylated and sialylated glycan at position 1098 along the MD trajectory.
Spike Protein Rigidity
The S protein was found to have both rigid or structured regions, that bind to the antibody strongly and specifically, and dynamic regions. The receptor-binding domain (RBD) and nearby regions are similarly flexible, which supports research showing marked differences in the peptide chain structure in both open and closed states.
On the other hand, the S2 domain that covers the fusion peptide has rigid protein structures, which may be necessary to keep this part of the molecule stable in the prefusion configuration.
![Epitopes identified from MD simulations and bioinformatics analyses. Accessibility scores from (A) ray analysis and (B) Fab rigid body docking are combined with (C) rigidity scores, all averaged over 4 × 1.93 µs of S protein MD simulations. Also included are (D) a sequence conservation score [31], and (E) BepiPred-2.0 epitope sequence-signature prediction. (F) Combined epitope score. (G) Binding sites of known neutralizing antibodies. Higher color intensity in A-F indicates a higher score and higher color intensity in G indicates sites binding to multiple different antibodies. Epitopes identified from MD simulations and bioinformatics analyses. Accessibility scores from (A) ray analysis and (B) Fab rigid body docking are combined with (C) rigidity scores, all averaged over 4 × 1.93 µs of S protein MD simulations. Also included are (D) a sequence conservation score [31], and (E) BepiPred-2.0 epitope sequence-signature prediction. (F) Combined epitope score. (G) Binding sites of known neutralizing antibodies. Higher color intensity in A-F indicates a higher score and higher color intensity in G indicates sites binding to multiple different antibodies.](https://www.news-medical.net/image-handler/picture/2020/7/ssCapture.jpg)
Epitopes identified from MD simulations and bioinformatics analyses. Accessibility scores from (A) ray analysis and (B) Fab rigid body docking are combined with (C) rigidity scores, all averaged over 4 × 1.93 µs of S protein MD simulations. Also included are (D) a sequence conservation score [31], and (E) BepiPred-2.0 epitope sequence-signature prediction. (F) Combined epitope score. (G) Binding sites of known neutralizing antibodies. Higher color intensity in A-F indicates a higher score and higher color intensity in G indicates sites binding to multiple different antibodies.
Spike Protein Conservation
The researchers stress the importance of developing vaccines that elicit antibodies against relatively stable epitopes to prevent escape mutations from emerging and allowing the new strains to replicate and becoming predominant. The current analysis found that S protein is highly conserved, and no variants are present in the current large databases for over half of the amino acids in this protein.
Predicting Immunogenicity
The researchers point out, “Conserved, rigid, and accessible regions present good candidates for binding of protein partners in general.” Using these features, they found 9 candidate epitopes, 4 of which are known and the remaining novel epitopes. All were in the head region of the spike protein, a structured portion of the molecule, unlike the flexible and inaccessible stalk region.
Agreement with Prior Research
A previous team of researchers found that the neutralizing antibody CR3022 bound to the S protein’s ectodomain, at an epitope beyond the ACE2 binding region. To achieve this binding, at least two of the three S protomers must be in the open configuration. This epitope is recovered in the current analysis, as well as epitopes that bind to other antibodies like CB6, H104, and S309.
Two of the other epitopes also match the RBD binding sites already reported for neutralizing antibodies, showing, in the researchers’ words, “that our epitope-identification methodology is robust.”
Glycosylation Sites Revealed
The study also confirmed and carried forward earlier research on the impact of different patterns of glycosylation on the accessibility of the protein. They found that whether mannose alone was included or the full glycan shield was considered, Fab was blocked effectively by 60% and 80%, respectively.
The researchers say that “even a light glycan coverage strongly reduces the antibody accessibility of the protein.”
Characteristics of Immunogenic Epitopes
They found that the first three epitopes had flexible loops as well as beta-folded strands. The next was on one alpha-helix flanked by another, lying on a beta-sheet of five strands, a very stable arrangement.
The next two are on the apical portion of the RBD on the S protein, mostly flexible loops that show conformational changes in the open and closed states. Others consist of long and short flexible loops or a stable helix.
Future Vaccine Design
The researchers sought to present the identified epitopes to maintain a robust immunogenic profile. They say that since the S protein is composed of distinct domains if these domains fold independently, they could conserve the epitopes and thus allow antibodies to be elicited by a vaccine. This could perhaps be served by suitably modifying the sequences.
The study concludes: “The approach we introduced in this paper could be extended to predict epitopes from an integrated analysis of diverse betacoronaviruses, with the ultimate aim of producing a universal vaccine that guarantees broad protection against the whole virus family.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources