The search for an effective antiviral against the COVID-19 scourge is continuing at high speed, given that second and third waves are expected to occur soon. Now, an intriguing new study out of the University of Wisconsin-Milwaukee reveals that numerous small molecule drugs like the proton pump inhibitors omeprazole and rabeprazole bind the important Mac1 domain on the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. The discovery, published on the preprint server bioRxiv*, could lead to the design of new drugs that bind this domain with equal affinity.
A direct-acting antiviral (DAA) is one that is a potent viral enzyme inhibitor, or a ligand that binds viral proteins at high affinity. These are being developed as a means to stem the spread of the virus. The only drug specifically approved against the virus so far is remdesivir, a prodrug that is converted to a nucleoside triphosphate. This acts to terminate the chain replication of the virus by inhibiting its RNA dependent RNA polymerase, bringing viral replication to a stop.
However, prior viral therapies have shown that it is better to use DAA cocktails of more than one antiviral, to avert the rapid onset of drug resistance. This lends urgency to the need to come up with more small molecules that can bind to multiple viral proteins so that they can be used to design new drugs as well as molecular probes that help delineate the biological role of these proteins in the viral lifecycle.
The Mac1 Domain
The RNA genome of this virus has approximately 29 kb, encoding four structural and 16 non-structural proteins (nsps), with 6 accessory proteins and other products as well. These are all potential DAA targets. Some are unique to this virus, including the 3’-5’ RNA proofreading exonuclease.
The viral genome has several open reading frames (ORFs), among which the ORF1ab encodes most of its nsps. The ORF1a protein is split into smaller fragments constituting the nsps 1 to 10, and the ORF1b into nsps 1 to 16. The current study focuses on the 945 amino acid viral nsp3, a protease that cleaves the three sites separating nsp1, nsp2, and nsp3.
In other coronaviruses, this enzyme is attached by two transmembrane helices to the lumen of the endoplasmic reticulum (ER) within the cell. Thus, most of it lies inside the cytoplasm. There are many domains on the N-terminal side of the transmembrane domains, including three macrodomains Mac1, Mac2, and Mac3.
Among these, the Mac1 domain is called the X domain and is considerably different from the nearest homolog in SARS-CoV. This domain binds ADP-ribose and is indeed capable of removing covalently attached ADP-ribose from proteins by its hydrolytic activity. This may be related, some research suggests, to the cytokine storm that is found to occur in severe COVID-19.
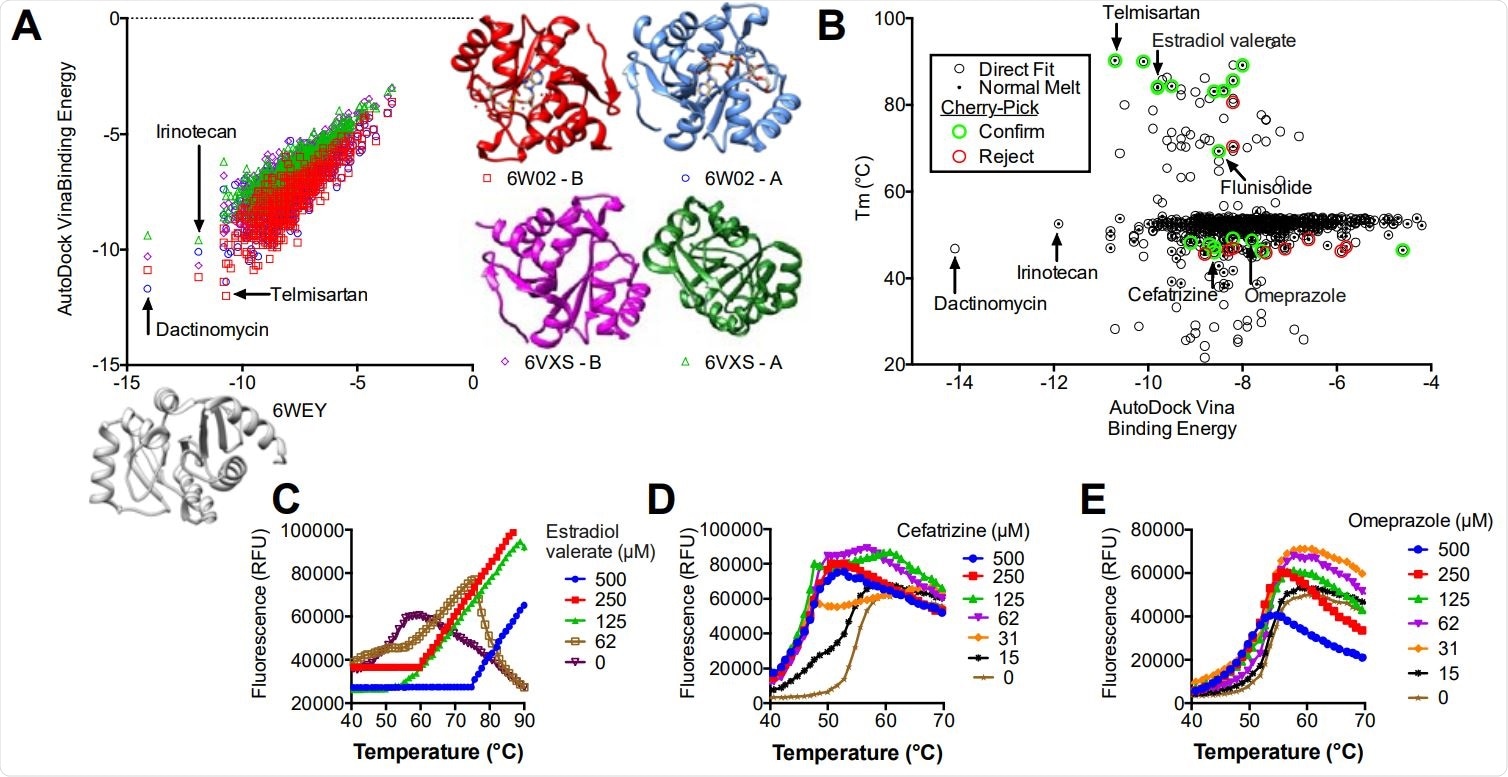
Virtual screening of the NIH clinical collection. (A) AutoDock Vina binding energies obtained for each compound after docking with PDB file 6WEY (x-axis) compare to binding energies when each compound was docked with BioAssembly A of 6W02 (circles) BioAssembly B of 6W02 (squares), BioAssembly A of 6VXS (triangles) and BioAssembly B of 6VXS (diamonds). (B) AutoDock Vina binding energies obtained for each compound after docking with PDB file 6WEY (y-axis) compared with the Tm derived using TSA-CRAFT (see Fig. 2). Compounds selected for follow-up (cherry pick) analysis are highlighted. (C-E) Concentration response analysis of three selected compounds (see text for details).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Study: Identifying Mac1 Ligands
The current study uses two high-throughput screening methods to identify potential Mac1 ligands from drug and drug-like compound libraries. These are differential scanning fluorimetry (DSF, aka the thermal shift) and a docking assay to find the compounds with the highest affinity for all structures. The researchers then tested the hits, or positive compounds, and found several molecules that performed well in both assays. Molecular models showed that these would probably bind in the Mac1 cleft where ADP binds.
Promising Hits Binding the ADP-Cleft
Of the various hits by the DSF assay, the ACE inhibitor telmisartan and the two steroids, estradiol valerate and the anti-inflammatory corticosteroid flunisolide were promising. Simultaneously, the virtual docking screen showed telmisartan, as well as other compounds such as the topoisomerase inhibitor irinotecan and the RNA synthesis inhibitor dactinomycin (actinomycin D).
On closer examination, some of the ligands that lowered the melting temperature in the DSF assay were from similar chemical compartments. Such ligands are thought to bind to unfolded protein structures and stabilize them. Some were from the lactam class of antibiotics, while others were benzimidazoles similar to telmisartan in their chemical structure.
For most of the hits, the binding energy was minimal when the compound docked near the ADP-binding site. This was true of the steroids, beta-lactams, and benzimidazoles, with the identified molecules occupying this cleft on Mac1. The larger the compounds, the higher the number of amino acids that come into contact, which in turn gives them higher binding energies.
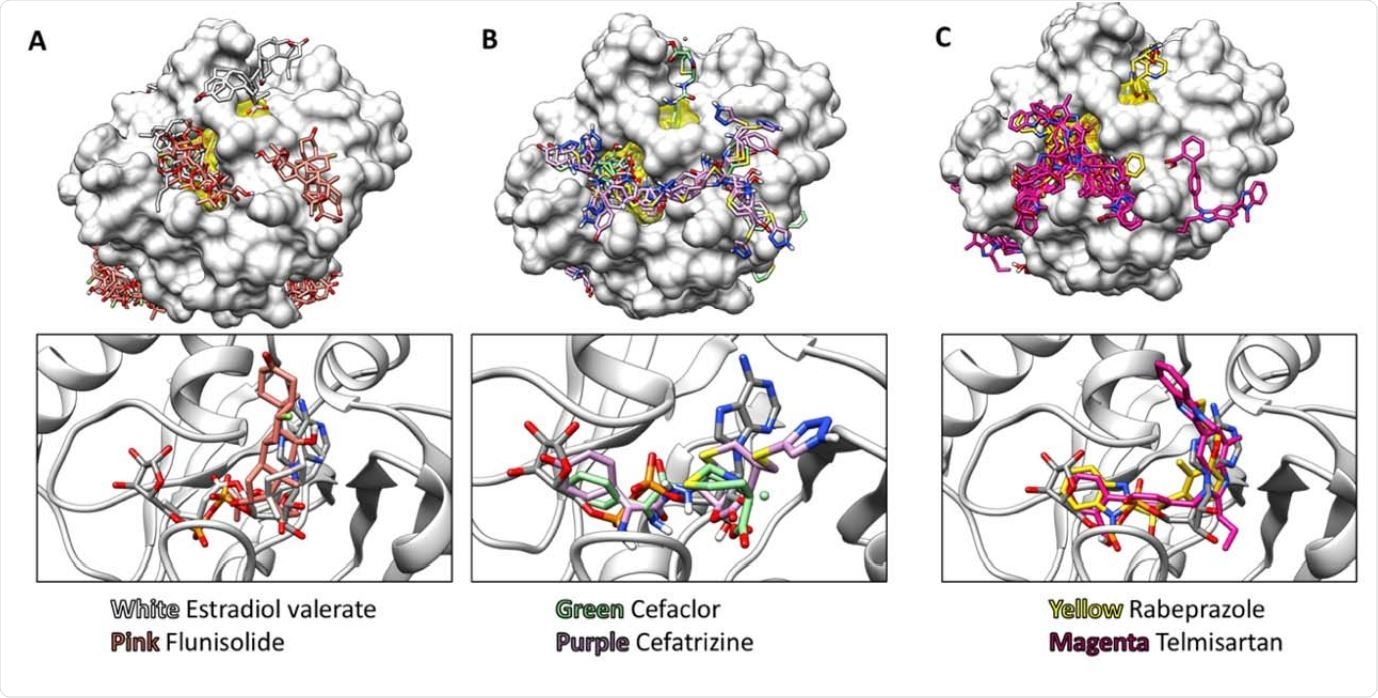
Representative Structures obtained using AutoDock Vina Virtual Screening. Top panels show the top 20 binding modes for selected compounds, and the bottom panels show the top binding mode for each compound compared to the ADP-ribose bound in PDB file 6W02. The ADP-ribose binding cleft is highlighted in yellow on the protein surfaces shown in the top panels. Results are shown for (A) two steroids, (B) two lactam antibiotics, and (C) two benzimidazoles in the NIH clinical collection. Structures are displayed using UCSF-Chimera.
Future Directions and Applications
The next step is to confirm that these are effective ligands, which would provide a foundation for a rational design for derivatives with higher affinity to this site. If so, this domain could act as an enzyme in the cell to remove ADP-ribose from antiviral proteins.
Despite the limitations of the methods used, the current study promoted these efforts by identifying compounds that bind with Mac1 in a similar way to ADP-ribose. The researchers point out that only those enzymes in the poly (ADP-ribose) polymerase (PARP) class that transfer a single ADP-ribose to an amino acid produce substrates that can be acted upon by Mac1.
Enzyme-based assays could be designed for this domain to help identify more potent inhibitors. Finding ligands that bind to Mac1 could, they say, guide “the nsp3 protease to cellular proteins to degrade, deubiquitinate, or catalyze the removal of Interferon-stimulated gene 15 proteins.” To test this hypothesis, molecules that bind this domain with high affinity need to be found. Hence the importance of the current study.
The fact that steroids were identified as Mac1 ligands may help explain the clear sex difference in the severity of COVID-19. The ACE inhibitor telmisartan could also be an antiviral compound. Much more work is required to examine how these ligands interact with Mac1. However, the other benzimidazoles could be verified using an independent assay method - rabeprazole, omeprazole, and esomeprazole. Their mechanism of action should be explored further – do these ligands bind to the protein in its unfolded state, or to a folded state but with a different conformation?
Further work will be necessary to see how these compounds act on infected cells, and whether they inhibit viral replication. The use of optimized probes can help screen larger libraries to find better probe candidates.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Virdi, R. S. et al. (2020). Discovery of Drug-like Ligands for the Mac1 Domain of SARS-CoV-2 Nsp3. bioRxiv preprint. doi: https://doi.org/10.1101/2020.07.06.190413. https://www.biorxiv.org/content/10.1101/2020.07.06.190413v1
- Peer reviewed and published scientific report.
Virdi, Rajdeep S., Robert V. Bavisotto, Nicholas C. Hopper, Nemanja Vuksanovic, Trevor R. Melkonian, Nicholas R. Silvaggi, and David N. Frick. 2020. “Discovery of Drug-like Ligands for the Mac1 Domain of SARS-CoV-2 Nsp3.” SLAS DISCOVERY: Advancing the Science of Drug Discovery, September, 247255522096042. https://doi.org/10.1177/2472555220960428. https://slas-discovery.org/article/S2472-5552(22)06649-7/fulltext.