The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which is currently at the heart of a pandemic that has cost the world more than 550,000 lives, 12 million infections, and uncounted years of productivity, is an RNA virus with the largest genome among all such viruses. This 30 kb size virus has caused many difficulties for scientists in search of a vaccine.
.jpg)
Novel Coronavirus SARS-CoV-2 This scanning electron microscope image shows SARS-CoV-2 (round gold objects) emerging from the surface of cells cultured in the lab. SARS-CoV-2, also known as 2019-nCoV, is the virus that causes COVID-19. The virus shown was isolated from a patient in the U.S. Image captured and colorized at NIAID's Rocky Mountain Laboratories (RML) in Hamilton, Montana. Credit: NIAID
The RNA in the viral genome is the template for translation of a complex array of enzymes that builds up the viral transcriptome – a set of subgenomic RNA sequences that encodes all the other components of the viral particle. This includes highly conserved bits of RNA that stay the same across strains and even families, probably because they are so important in viral replication. These structural elements with high functionality have been little explored in the SARS-CoV-2.
Previous reports have suggested that this virus also forms genome-scale ordered RNA structures (GORS) like other coronaviruses. This feature confers fitness and persistence. Since RNA structure is essential for viral function and for higher-order compaction, the current study focuses on this aspect of the viral genome.
Comparing RNA Secondary Structures
In a new study out of Yale University and published on the preprint server bioRxiv* researchers assessed the folding stability of the RNA genome of this virus in comparison to other known systems, to explore the biological contributions made by these structural features. Despite the size, they aimed to produce a broad landscape of the overall as well as detailed structure and organization of the virus, which would help to identify those regions that had the most potential for regulating its life cycle.
The study conducted a comparative structural analysis of SARS-CoV-2 genomic RNA as well as of the previously known most highly structured RNA virus, the hepatitis C virus (HCV), the West Nile virus which is considered to be lacking overall RNA structure, and a set of human mRNAs which do not have internal structure.
They use what is called a Z-score to assess the tendency to form stable RNA structures. Negative Z-score values suggest the presence of very stable secondary structures in the RNA. This was -0.35 (median) for the human mRNAs, and -0.2 for the West Nile virus, suggesting low base pairing. On the other hand, the Z-scores for the HCV genome were almost always -1 throughout, showing that the tendency to form base pairs was high throughout the genome.
The SARS-CoV-2 shows a much more negative Z score distribution, showing its much higher tendency towards stable base pairings leading to the formation of more secondary structures, much more than any other RNA analyzed so far and not explicable as a coincidence. The median value of -1.5 shows it is likely to form a large and intricate pattern of functional secondary structures in coding and non-coding regions of its functional domains.
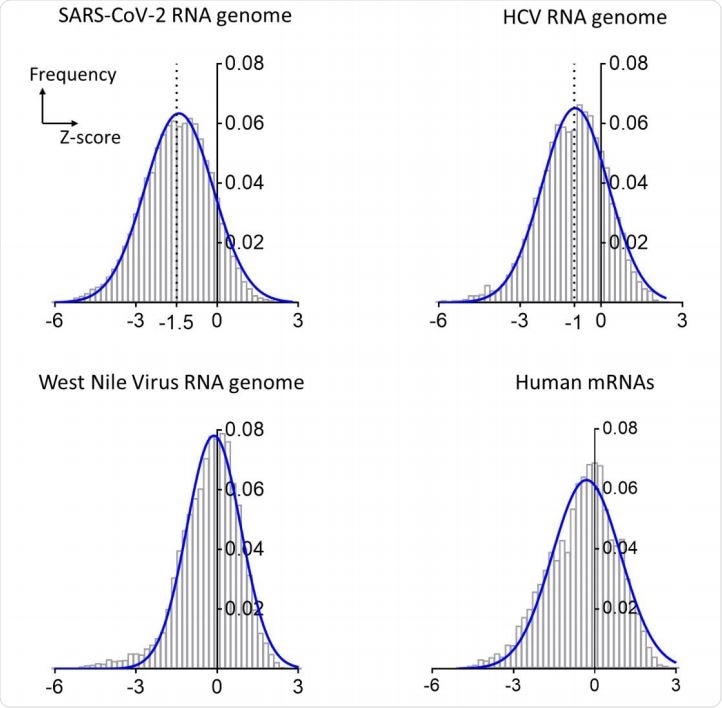
Distributions of Z-scores for the RNA genomes of SARS-CoV-2, HCV and West Nile viruses and a composite of human mRNAs. The bar plots are frequency distributions (y-axis) of free-energy Z-scores (x-axis) calculated in sliding windows tiling each RNA. Each histogram is overlaid with a Gaussian (normal distribution) fit represented by a solid blue curve.
The study shows that the RNA folding stability of this genome is twice that of the HCV, hitherto the hallmark of stable secondary structure. This complex structure is biologically very significant.
Developing Platforms for Base Pair Quantification
They constructed a workflow and the tools to decipher the base pair content from the secondary structure of any long RNA as well as to pick up any structural definition in a transcript containing kb of base pairs. Using this, they identified the highly complex structural regions of RNA, so as to compare predicted structures among the domains of this large RNA.
They saw that the genome was mostly folded into stable and separate repeating structural elements or motifs, with the base pair content (BPC) being about 61% on average, which is in keeping with the Z-score. There are abundant stable secondary and tertiary well-defined structural regions within the genome.
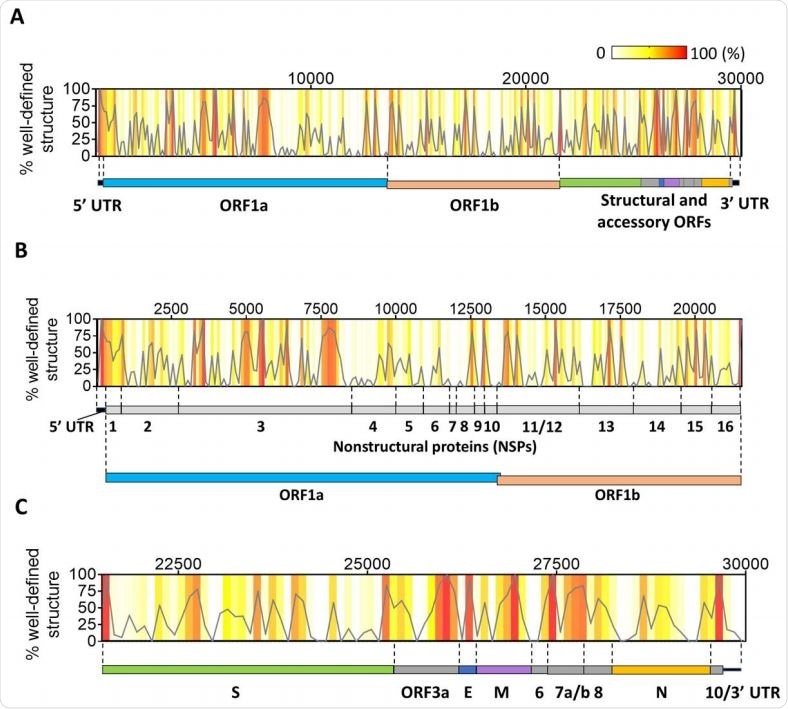
Distribution of well-defined RNA structures across the SARS-CoV-2 genome. (A) The percentage of nucleotides in well-defined structured regions (high BPC/low Shannon) was calculated in 100-nt bins tiling the genome and is plotted as a function of the genomic coordinate (gray curve). Individual percentages of each genomic bin are also represented as a heatmap in the same graph (color legend on the top right-hand corner). A scheme representing the genomic divisions of SARS-CoV-2 is shown next to the plot to guide the location of structured regions. (B) An expanded view of the initial two-thirds of the genome from the graph in (A) is shown along with the genomic divisions of this region (UTR + ORF1ab and corresponding NSP divisions). (C) The downstream third of the genome is expanded from the graph in (A) to zoom in on individual structural and accessory ORFs in this region.
Specific Regions of Highly Structured RNA
The result was that they found that both the untranslated regions, at the 5’ and 3’ end, had a high level of structural content, at over 60% and 40% respectively, which can be explained as the presence of RNA regulatory elements responsible for viral replication and translation.
The ORF1ab that forms the upstream two-thirds of the genome has many foci of RNA structures non-uniformly distributed along the genome, with the Novel SH2-containing Protein (nsp) 1 segment, that is the most highly structured part, containing 56% of its nucleotides in clearly defined structures. In fact, the upstream part of the nsp1 segment is part of a large module that forms in conjunction with the 5’ UTR, so that the upstream regulatory elements of the genome seem to overlap with the ORF considerably.
Other nsps form clusters of structural elements that are either confined within their respective domain limits or, as with nsp 4 to 6, 8 to 10, and 14-16, they may form structures that spill over the borders of the domain. This may mean that these RNA forms modules which are not always functional units. However, nsps 7 and 11 have no structured regions at all.
They also found a much higher number of structured regions among the open reading frames (ORFs), which code for structural and accessory proteins, and form the downstream one-third of the genome.
Genomic vs. Subgenomic Structures
They also found that these ORF structures can shift depending on whether they are in a genome or a subgenomic fragment. For instance, they examined the nucleocapsid ORF, the most plentifully occurring subgenomic RNA that is found one order of magnitude more abundantly than any other. Because of the differences in the base-pairing potential when the N ORF is in the genome or separate from it, secondary structures that are formed in these contexts are different, and presumably, carry out different functions altogether. This could allow different levels of RNA stability, processing, and molecular functioning.
The Why and the Future of RNA Structural Studies
The reason for such massive structuring could be protective, as this does not allow cellular nucleases to gain access to it and destroy it, allowing infection to continue. It may also enable immune evasion by minimizing the recognition patterns that can be picked up by pattern recognition receptors in the host cell. Thirdly, it allows distant elements of the large genome to interact by bringing them close to each other.
The study has thus brought out results that will provoke greater exploration of the complex ways in which viruses infect their hosts and how host responses occur. The uncovering of viral genomic structure could be immensely useful in finding new drug targets to aim at in the fight to develop a treatment for COVID-19.
The researchers conclude, “The conclusions reported in this study provide a foundation for structure-function hypotheses in SARS-CoV-2 biology, and in turn, may guide the 3D structural characterization of potential RNA drug targets for COVID-19 therapeutics.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources