Researchers from the Venezuelan Institute for Scientific Research and the Australian National University untangle the mutational and recombinational history of the SARS-2 lineage of betacoronaviruses and their metagenomes with the use of published genomic sequences. Their findings are currently available on the bioRxiv* preprint server.
An ongoing pandemic of coronavirus disease (COVID-19) caused by a novel betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains a substantial global health issue with significant repercussions for societies around the world.
Nonetheless, coronaviruses have been recognized as pathogenic agents since the 1960s, infecting humans (with resulting respiratory or gastrointestinal infections), but also a variety of animal species – including mammals and birds.
Coronaviridae can be classified into four genera: Alphacoronavirus, Betaoronavirus, Deltaoronavirus, and Gammacoronavirus. Betacoronavirus genus is further separated into five subgenera (Sarbecovirus, Hibecovirus, Embecovirus, Merbecovirus, and Nobecovirus).
We are already well aware that COVID-19 is a zoonosis; still, while trying to establish from where SARS-CoV-2 exactly emerged, there have been conflicting claims where either bats or pangolins are implicated as intermediary hosts. What we do know is that all of them are subgenus sarbecoviruses of the genus Betacoronavirus.
Dr. Eduardo Rodríguez-Román, a biologist and virologist from the Venezuelan Institute for Scientific Research, together with Professor Adrian J Gibbs, virologist, and evolutionary biologist from the Australian National University, resolved some of the aforementioned controversies and unveiled a fascinating shared betacoronavirus region.
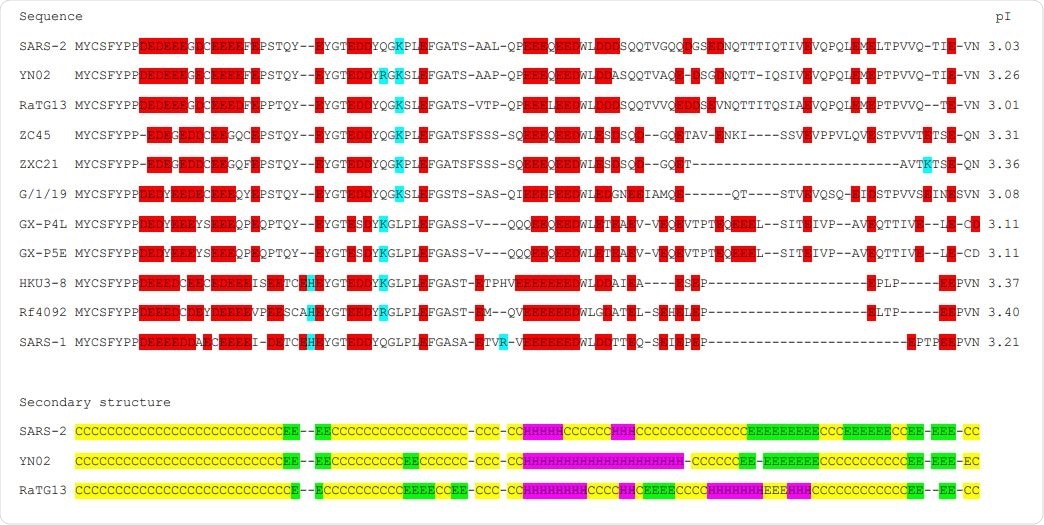
Comparison of the DUF3655 region of the eleven betacoronaviruses analysed in this study

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Exposing full-length genomic sequences
"We have discombobulated the recombinational and mutational history of the SARS-2 lineage of betacoronaviruses and their metagenomes using the published genomic sequences", study authors summarize their research approach.
More specifically, in mid-May 2020, they have conducted a similar BLAST search of the Genbank nucleotide sequence databases using the SARS-CoV-2 Wuhan-Hu-1 sequence as a search query. This resulted in the identification of over one hundred related full-length genomic sequences.
These researchers have also compared the concatenate sequences (concats) directly in pairs – not only to pinpoint any regions that show abnormal evolution but also to confirm the distinct recombination map patterns. Other sequence comparisons (such as DnDscan) were also pursued.
Clear evidence of recombination
"We have shown that the 5' third of SARS-2 betacoronavirus genome is largely free of recombinant regions, whereas the remainder is a mélange of recombinant regions from various 'parental' genomes", study authors explain their findings.
In any case, all of them showed unambiguous evidence of recombination, and most events involved the 3' half of the genomes (i.e., 'back end' of the respective RNA strain).
A distinctive pair of recombinant regions were found in the SARS-2 crown group, most closely related to the homologous region of the SARS-1 lineage bat virus known as Rf4092. Moreover, a phylogeny calculated from the 5' n-rec region of the eleven concats reveals that the SARS-2 lineage has basal branches of viruses that were isolated from pangolins.
Most of the SARS-2 concats (especially its 5 prime end) were closest to the homologous regions of YN02 bat coronavirus; however, the intact concats of SARS-2 and RaTG13 were more distant but complete.
"Phylogeny analysis showed that SARS-CoV-2 diverged from RmYN02 at least 26 years ago, and both diverged from RaTG13 at least 37 years ago; recombinant regions specific to these three viruses provided no additional information as they matched no other Genbank sequences closely", study authors add.
The significance of DUF3655 region
Simple pairwise genome comparisons revealed three regions with most non-synonymous changes – the DUF3655 region of the nsp3, ORF 8 gene, and the S gene. Recombinational changes most likely caused differences in the last two of those regions; however, differences in the DUF3655 region may have arisen due to a selection process.
The same DUF3655 region thus far successfully evaded virological, pharmaceutical and medical scrutiny, although it seems to be a superb target for developing effective treatment approaches for sarbecoviruses.
"We suggest that it is probably involved in a unique rate-limiting step of the coronavirus replicative cycle, and may make coronavirus infections susceptible to drugs, like chloroquine, that increase cellular pH," hypothesize study authors.
In any case, the DUF3655 region warrants more attention, since repetitive acidic amino acids can be observed in similar parts of the genomes. Additional research will be needed in order to fully utilize this potentially novel target for pharmaceutical intervention.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources