Even as COVID-19 continues to spread around the world, researchers are focusing on vaccine and antiviral development. The target of much of this study is the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a trimeric component of the viral particle that is responsible for attachment to the host receptor, the angiotensin-converting enzyme 2 (ACE2). Now, a new study by researchers at Janssen Vaccines & Prevention BV and Leiden University and published on the preprint server bioRxiv* uses cryoEM to reveal the structure of this trimer, in order to support both vaccine development and diagnostic testing based on antibody production.
The S protein is a large glycoprotein composed of two subunits, mediating both host cell receptor binding and membrane fusion of the virus and the host cell to enable viral entry and infection of the host cell. The S1 subunit has two separate parts, an N-terminal domain (NTD) and a receptor-binding domain (RBD).
.jpg)
3D Illustration of SARS-CoV-2 Structure Diagram. Image Credit: Orpheus FX / Shutterstock O By

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
At the S1/S2 interface, there is a furin-like protease that is required for the cleavage of the protein, which can also be carried out by the action of TMPRSS2, a highly conserved site just before the S2 fusion peptide. The pre-fusion S protein has its RBD alternately in the up and down or open and closed configurations.
In the former, the binding site of the receptor is transiently exposed to the host cell receptor, or to any antibody. Because of this change in conformations that intrinsically characterizes the protein, the S protein is, like other class I fusion proteins, a naturally metastable protein.
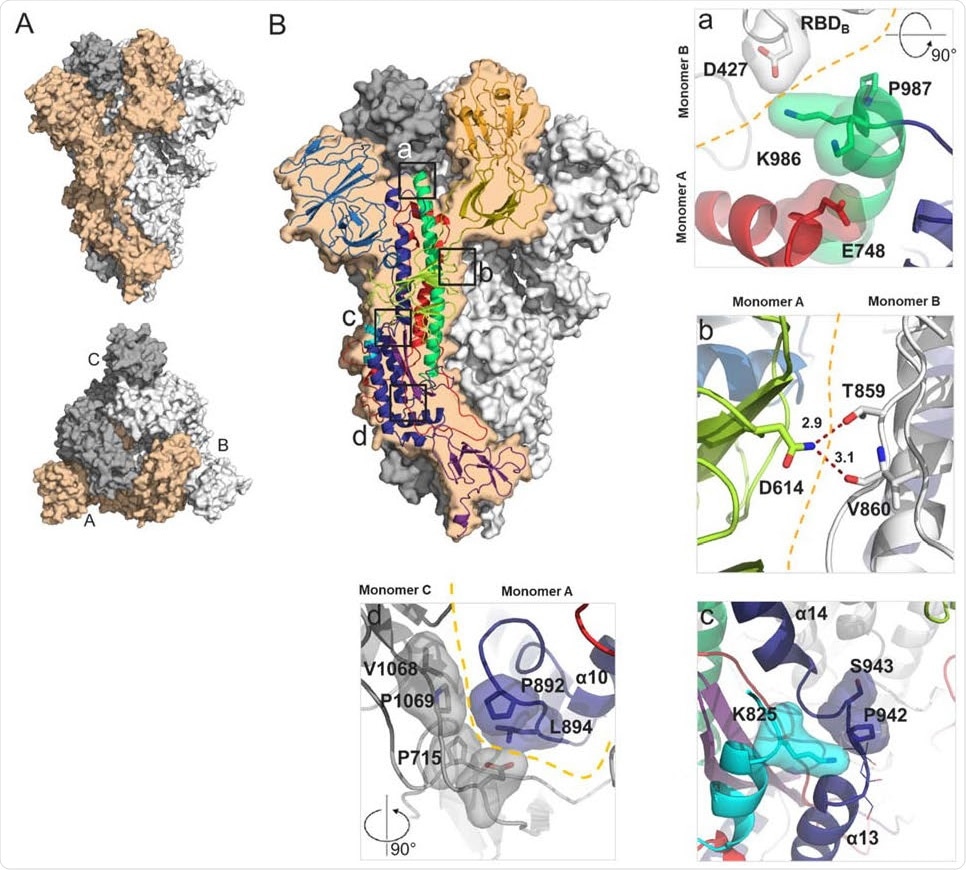
Structural characterization of S-closed. (A) Cryo-EM structure of the most abundant trimer class of S-closed-Fd – a closed S protein trimer. Monomers are colored in light orange, white and grey and the spike is shown from the side (upper panel) and from the top (lower panel) view. (B) Each of the four single point mutations introduced in S-closed-Fd shown in detail. Domains of the new structure are colored according to the same color code as used in Fig S1. Boundary between monomers has been additionally indicated with orange dashed line when applicable. Two possible rotamers of K986 are shown as one or the other could not be definitively assigned based on the density.
Targeting the Prefusion S Protein
The prefusion form of the S protein contains the epitopes to which neutralizing antibodies bind, and is, therefore, a prime target for immunogen development. For this to occur, it is necessary to stabilize the prefusion protein, as thereby more recombinant glycoproteins on the viral particles are expressed, stimulating an immune response.
One way to stabilize this protein is by stabilizing the hinge loop that lies just before the central helix. In the earlier SARS-CoV and MERS-CoV, this was accomplished by mutating two successive amino acids to proline in the S2 subunit in the region lying between the central helix and the heptad repeat (HR) 1.
This has been carried over into the SARS-CoV-2 S protein as well (SARS-CoV-2 S-2P). On the other hand, this mutated S protein is unstable. The current study describes ways to enhance its stability.
Another aspect to be considered is that neutralizing antibodies have the highest potency when they bind to the RBD epitopes in the closed or downstate. This means the prefusion protein must be stabilized in this conformation to obtain a stronger immunogen.
Stabilizing the S Protein in Closed Conformation
The instability of the S protein thus does not improve entirely even with the introduction of the double proline in the hinge loop. The current study, based on structure-based design, identified novel stabilizing mutations in S1 and S2 domains.
The researchers demonstrate that along with enhanced trimer expression by the individual proline mutations, the K986P variant decreases the interaction with the S1 head. This causes the RBD to be in the upstate, as assessed by increased binding with both the ACE2 and the CR3022 antibody. When the second stabilizing mutation, namely, V987P, is in place, this effect is partly compensated.
The researchers show the effects of two additional groups of substitutions. The first is in the S2 heptad repeat (HR) 1 region, which is radically affected by fusion. In the second, proline and glycine take part in several loop mutations to stabilize the trimer, increasing the yield. These S2 mutations probably help with protein folding required for the conformational change that triggers fusion. Thus, the expression of the prefusion S protein increases.
They then home in on four single-point mutations (D614N, A892P, A942P, and V987P) that cause the S protein to remain in a closed stable conformation, but with a 6.4 times higher expression, stability to heat and to freeze-thaw cycles, and antigenic properties similar to that of a closed trimer.
Combining Mutations for Optimum Effectiveness
When several of these mutations were combined, the researchers obtained a very stable S-closed trimer with higher glycoprotein expression. They included both D614N and A892P, both of which stabilized the S protein, and A942P, which increased the yield of the protein.
Combining these with the prolines in the hinge loop gave a quadruple mutant (D614N+A892P+A942P+V987P), and a quintuple mutant (all these plus K986P). Both produced a fivefold increase in yields, relative to the S-2P variant, but the latter had increased ACE2 binding. When the heterologous trimerization domain was deleted, the protein continued to be stable, and in fact, the protein yield went up 6.4 times relative to the S-2P variant, even though soluble S protein designs find this foldon domain to be essential.
Why Target Closed Spike Protein?
The importance of targeting the spike protein with the closed RBD conformation is that this is how the virus is during the process of transmission. Thus, antibodies that target the virus in this state are likely to be more protective against infection. Secondly, in this state, the surface is more conserved, and therefore more cross-reactive antibodies can be induced.
The researchers sum up: “A stable closed S trimer with minimal non-exposed mutations and without a foldon that shows a significant increase in expression levels may advance the development of novel (subunit) vaccine immunogens and further improve genetic vaccines, diagnostics or isolation of antibodies.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Juraszek, J. et al. (2020). Stabilizing the Closed SARS-CoV-2 Spike Trimer. bioRxiv preprint doi: https://doi.org/10.1101/2020.07.10.197814. https://www.biorxiv.org/content/10.1101/2020.07.10.197814v1
- Peer reviewed and published scientific report.
Juraszek, Jarek, Lucy Rutten, Sven Blokland, Pascale Bouchier, Richard Voorzaat, Tina Ritschel, Mark J. G. Bakkers, Ludovic L. R. Renault, and Johannes P. M. Langedijk. 2021. “Stabilizing the Closed SARS-CoV-2 Spike Trimer.” Nature Communications 12 (1). https://doi.org/10.1038/s41467-020-20321-x. https://www.nature.com/articles/s41467-020-20321-x.