The current COVID-19 pandemic is one of the most potentially devastating threats presented in the form of a disease, to the world. As scientists work to understand the virus and find a vaccine, the transcriptional landscape of the host cell following infection has come under intense scrutiny.
Now, a new study by researchers from the Icahn School of Medicine at Mount Sinai and Weill Cornell Medical College in the USA, that is published on the preprint server bioRxiv* in July 2020 describes how compounds that restore the normal transcriptional conditions following infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be used to reduce viral load without neutralizing the virus entry into the host cell.
The authors say: “Ideally, in the face of a pandemic, we would be able to deploy a pan-antiviral that universally targets a transcriptional footprint common to all viruses.”
Issues with Pan-Specific Antivirals
There are some challenges to this idea. One is that all viruses being obligate intracellular parasites, and incomplete organisms, they rely almost entirely on host cell machinery to replicate and amplify themselves and to spread. Thus, any drug which antagonizes these processes acts against the host cells themselves.
Secondly, every virus affecting human cells suppresses one or more components of the antiviral response automatically set in motion by the cell. Thus each virus produces a different kind of transcriptional landscape. The nearest approach to the pan-virus solution proposed above is the development of antivirals specifically targeting a family of viruses, such as those which antagonize orthomyxoviruses.
However, no antiviral broadly antagonizes all coronaviruses. As a result, researchers have turned to the repurposing of already approved drugs which have already been well-studied for toxicity and drug kinetics, and thus can be immediately used if they show any evidence of benefit.
Most efforts in this area rely on computational modeling to predict efficacy, or in vitro screening of drugs. Drug screens in vitro have not been too successful since something as basic as the cell chosen to express the viral antigen can change the way the drug reacts with it.
The Study: Finding Drugs that Oppose Viral-Induced Transcriptional Signatures
The current research focuses on using sequencing data from a range of cell types and models. One such database is the library of integrated network-based cellular signatures (LINCS).
The LINCS is an extensive collection of characteristic molecular profiles resulting from transcription, translation, replication, metabolism, and various other processes that continuously occur within a given cell. The current study aims to use this as a source from which compounds can be identified that oppose or simulate the changes in gene expression caused by any given disturbance.
For instance, after a viral infection, the particular pattern of changes in gene expression can be identified and used to find those alterations that would need to be restored to bring the cell back to normal. This can also be used with drug treatment. A systematic application of this would find antivirals that act by restoring homeostasis to the infected cell. Prior research by the current team found a unique transcriptional footprint in lung cultures infected with the SARS-CoV-2. This was followed by RNA sequencing based on the RNA from these cells, to create various collections of gene expression signatures unique for this infection.
This was then used to interrogate the LINCS L1000 dataset, which contains the transcriptional reaction to the administration of more than 20,000 bioactive compounds to human cells in culture over a range of times and concentrations. This includes more than 1,000 drugs already approved by the FDA.
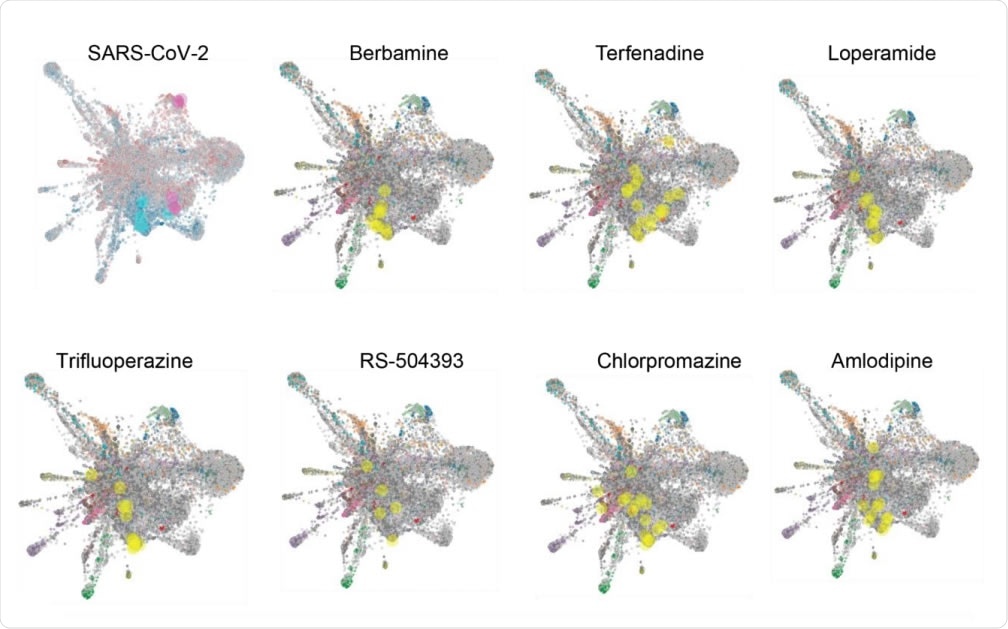
L1000FWD analysis of SARS-CoV-2 and candidate drug transcriptional space. A) L1000FWD analysis projecting the SARS-COV-2 uninfected vs. infected A549-ACE2 RNAseq signature onto the L1000FWD space (a) where red represents mimicking signatures and blue represents reversing signatures. Top 5 mimickers are highlighted in pink and top 5 reversers are highlighted in cyan. Projection of 7 reversing drugs: terfenadine, loperamide, berbamine, amlodipine, trifluoperazine, RS-504393, and chlorpromazine. Here signatures are colored by their known mechanisms of action where the signatures for each drug are highlighted in yellow.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Targeting Uniquely Affected Gene Region
The researchers were able to find transcriptional signatures that were inversely related between any given compound and viral infection. Specifically, they found a high-dimensional region in the gene expression space that is unique to this virus.
They found that 50 genes in this region were downregulated, or expressed at a lower level, in SARS-CoV-2 infected cells, but upregulated by eight drugs from this platform. This is supported by another drug screen study for compounds that inhibit this infection in Vero E6 cells, which also identified several of the same compounds.
The identified drugs were validated in many cell-based assays, using Vero cells, A549 cells, and human organoids. They found that when treated one hour before infection, seven of these drugs completely prevented the detection of the viral nucleocapsid. Viral replication was stopped or severely reduced by 2-3 orders of magnitude. However, they could not find any impact on the viral entry in response to any of these compounds.
Target: Cholesterol Biosynthesis Pathway
They then exposed cells expressing ACE2 to the drugs, with or without the virus, and assessed gene expression over the whole genome using RNA sequencing. This showed that the transcriptional signature is that of the cholesterol biosynthesis pathway. Perturbations in this pathway disrupt the viral lifecycle, since, for instance, extensive intracellular membranes (made up of cholesterol to a large extent) are required for the replication of SARS-CoV-2.
Four Candidates
This led to the final identification of four promising drug candidates, namely, amlodipine, loperamide, terfenadine, and berbamine. Amlodipine is an antihypertensive, while loperamide is an antidiarrheal drug. Terfenadine used to be employed as an antiallergic but was withdrawn for its cardiac toxicity. Amlodipine and terfenadine were found to upregulate the type 1 interferon signaling pathway. Berbamine is an experimental drug that regulates signal transduction and is a natural compound from the plant Berberis amurensi.
Each of these has an independent mechanism of inhibition of viral replication and suppressed viral transcription by three orders of magnitude.
Altogether, these suggest that such small molecules could be a genuinely novel approach to antiviral therapies. This should encourage in vivo testing to find out their therapeutic effectiveness in clinical situations. Even if these drugs are not ultimately found to be useful, these findings could help understand virus-host interactions in greater detail.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.