Researchers from the University of Bristol, Oxford Brookes University and the University of California San Diego neatly demonstrated how the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) exhibits high affinity for nicotinic acetylcholine receptors (nAChRs), with significant implications for coronavirus disease (COVID-19) pathology and infectivity. Their findings are published on the bioRxiv* preprint server.
The ongoing COVID-19 pandemic, caused by SARS-CoV-2, remains a substantial threat to global health, the international economy and society as a whole. Several major risk factors for COVID-19 have been identified – namely, age, diabetes, hypertension, and heart disease.
Recently, given the seemingly low prevalence of smokers among hospitalized patients, it was suggested that nicotine might provide some protection in mitigating COVID-19, which was dubbed the 'protection' hypothesis.
More specifically, based on the early observations where smoking prevalence in hospitalized COVID-19 patients was lower than expected, certain studies suggested a role for nAChRs in the pathophysiology of COVID-19 through a direct interaction between these receptors and the viral spike glycoprotein (S-protein).
This suggestion was primarily based on the fact that the S-protein from SARS-CoV-2 harbors a sequence motif related to known nAChR antagonists and may interact with nAChRs. Consequently, such interactions may be then involved in pathology and infectivity, which is a notion known as 'nicotinic hypothesis.'
Furthermore, it was also proposed that COVID-19 might be controlled or alleviated by the use of nicotine if this compound can sterically or allosterically compete with the virus for binding to nAChRs.
In this novel study, the researchers used molecular simulation to examine the nicotinic hypothesis – primarily by appraising whether the SARS-CoV-2 S-protein can stably bind to nAChRs via the Y674-R685 region (i.e., a viral portion with the highest affinity to these receptors).
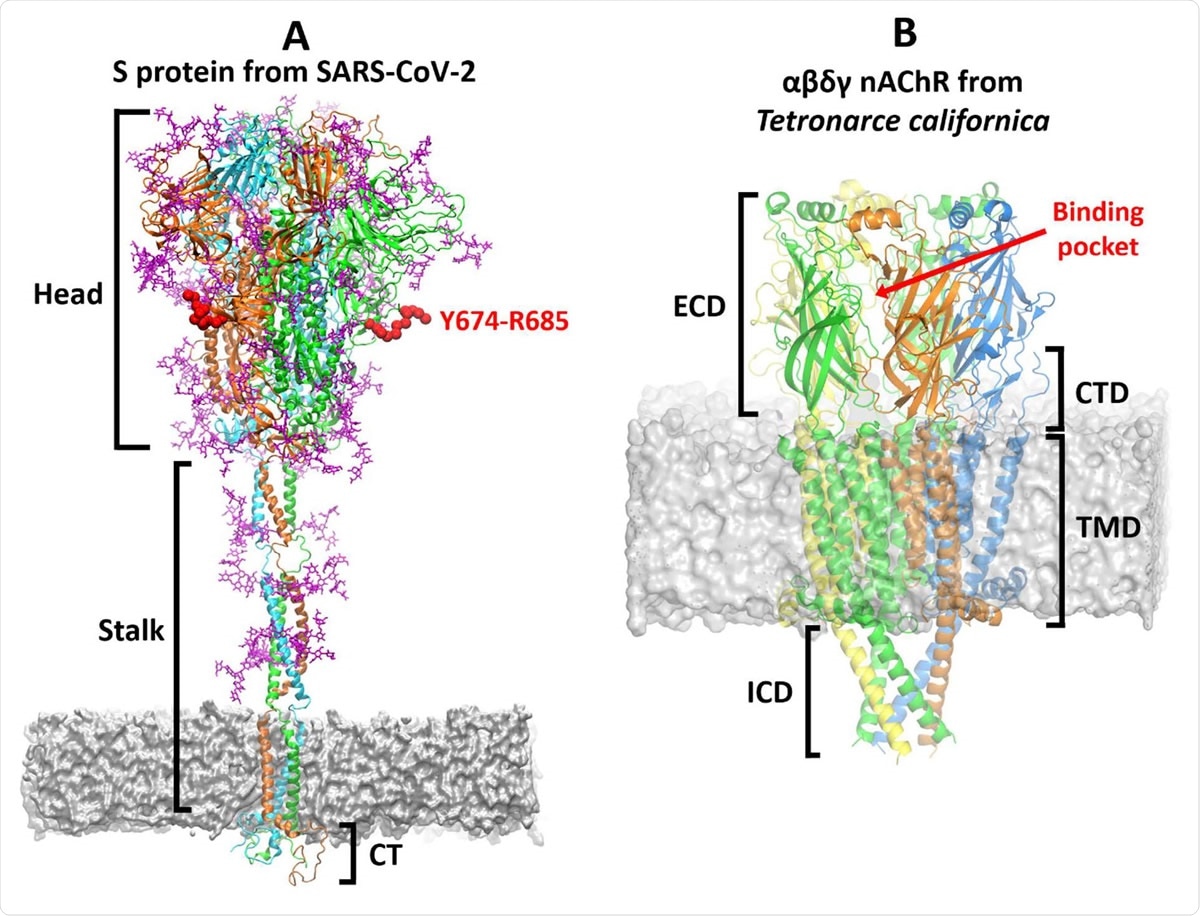
Overview of the three-dimensional structures of the S protein from SARS-CoV-2 and the αβγδ nAChR from Tetronarce californica. (A) The model for the complete, fully glycosylated, SARS-CoV-2 S protein represents the closed state of the protein, after furin cleavage.17 The S protein is a homotrimer:20 each monomer is shown in a different colour, namely green, cyan and orange, with glycans depicted in pink. Each monomer is formed by three domains: head, stalk and cytoplasmic tail (CT).20 The Y674-R685 region is shown in red. In MD simulations of the glycosylated SARS-CoV-2 S protein,17 Y674-R685 is accessible, being only weakly shielded by the glycans (Figure S4) and also shows high flexibility (Figure S5). (B) The cryoEM structure of the muscle-type receptor from Tetronarce californica (PDB code: 6UWZ). 18 This receptor is a heteropentamer formed of two α (green), one β (blue), one δ (yellow), and one γ (orange) subunits. Each monomer is formed by four domains:14-16 extracellular (ECD), transmembrane (TMD), intracellular (ICD) and C-terminal domain (CTD). The agonist binding site is located in the ECDs at the interface between two neighbouring subunits.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Structural modeling and molecular mechanics
This paper entailed state-of-the-art experimental procedures to explore the binding of the Y674-R685 loop of the SARS-CoV-2 S-protein to three nAChRs – namely the human α4β2 and α7 subtypes, and the muscle-like αβγδ receptor from Tetronarce californica (i.e., a species of electric ray).
Structural models of the three SARS-CoV-2 S-peptide–nAChR complexes were constructed based on the cryo-electron microscopy structure of the αβγδ receptor with bungarotoxin. The latter is a neurotoxin that acts as a nAChR antagonist, directly competing with acetylcholine (i.e., a neurotransmitter that binds to nicotinic receptors).
Moreover, a molecular mechanics Poisson–Boltzmann surface area approach was employed to estimate the free energy of binding of the S-protein to the different receptors. This is an efficient and useful method to determine binding free energies, which is widely used to study protein-ligand interactions in medicinal chemistry and drug design.
Finally, in silico alanine-scanning mutagenesis was carried out in order to pinpoint essential residues (referred by the authors to as 'hotspots') which drive all peptide-receptor associations.
Verified interaction of SARS-CoV-2 with nicotinic acetylcholine receptors
In a nutshell, the findings reported in this study support the hypothesis that the SARS-CoV-2 S-protein can indeed interact with nAChRs. More specifically, the results indicate that the Y674-R685 region from the S-protein shows a significant affinity for nAChRs in general, with the highest affinity for the muscle-like receptor.
"Our calculations indicate stable binding of the S-protein to these receptors through a region adjacent to the furin cleavage site and corresponding to the Y674-R685 loop", study authors explain their findings. "They also show apparent subtype-specific interactions, with the highest affinity for the muscle-type αβγδ receptor", they add.
Of note, furin cleavage site has many implications for the viral life cycle. Additionally, the region in the S-protein that is responsible for binding to nAChRs shares high sequence similarity with neurotoxins known to be nAChRs antagonists.
"Analyses of the simulations of the full-length S protein show that the Y674-R685 region is accessible for binding, and suggest a potential binding orientation of the S protein with nAChRs", study authors further explain.
Finally, modeling studies of the interaction between the full-length S-protein and nAChRs show that association is achievable with the proteins in a non-parallel orientation to one another, which is a significant observation for further research endeavors in this field.
Smoking cessation agents as drugs for COVID-19?
We already know that COVID-19 can be responsible for a wide array of respiratory, muscular, and neurological symptoms. Hence, the interactions predicted in this study may be relevant for understanding the pathophysiology of this disease.
"If nicotine does indeed prove to have any clinical value, it would likely be due to interfering with the association with nAChRs," study authors conclude in their bioRxiv paper.
And if that is indeed the case, nicotine analogs (also referred to as smoking cessation agents) –such as varenicline, cytisine, and cytisine derivatives – could also find their place in treating patients with (potentially severe) COVID-19.
Taking into account these promising results, further mutational and structural studies will be required to test the importance of SARS-CoV-2 S-protein interactions with nAChRs, with its potential relevance to pathology and infectivity of COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Oliveira, A.S.F. et al. (2020). Simulations support the interaction of the SARS-CoV-2 spike protein with nicotinic acetylcholine receptors and suggest subtype specificity. bioRxiv. https://doi.org/10.1101/2020.07.16.206680.
- Peer reviewed and published scientific report.
Oliveira, A. Sofia F., Amaurys Avila Ibarra, Isabel Bermudez, Lorenzo Casalino, Zied Gaieb, Deborah K. Shoemark, Timothy Gallagher, Richard B. Sessions, Rommie E. Amaro, and Adrian J. Mulholland. 2021. “A Potential Interaction between the SARS-CoV-2 Spike Protein and Nicotinic Acetylcholine Receptors.” Biophysical Journal 120 (6): 983–93. https://doi.org/10.1016/j.bpj.2021.01.037. https://www.cell.com/biophysj/fulltext/S0006-3495(21)00146-6.