The pandemic of COVID-19 has spread rapidly and extensively, requiring urgent and intensive research to develop a vaccine as well as to design an antiviral preventive or therapeutic. These efforts require a complete understanding of viral structure.
Now, a new study by an international team of researchers and published on the preprint server bioRxiv* in July 2020 shows how this can be advanced at a greater speed by combining experimental results with homology models obtained by high-throughput pipelines. This will help produce new hypotheses that will show novel druggable targets for effective drug development.
The current study presents over 870 models of the structure of various proteins making up the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The sequencing and 3D data come from the latest entries in the protein database, which resemble any of the viral proteins by sequence. The researchers aligned the viral protein sequences to those of all 3D structures available in the PDB. They found that almost all the structures were those of viral proteins, but sometimes they were complexes of viral and host proteins. In a small number, they were of host proteins, indicating the virus was mimicking them.
These structures were then added to using prediction software to model the structure of over 32,700 proteins, and the whole set was then methodically explored to understand the features of these structures. They found only six proteins which showed evidence of binding to other proteins, structurally. These comprise two teams of three proteins each.
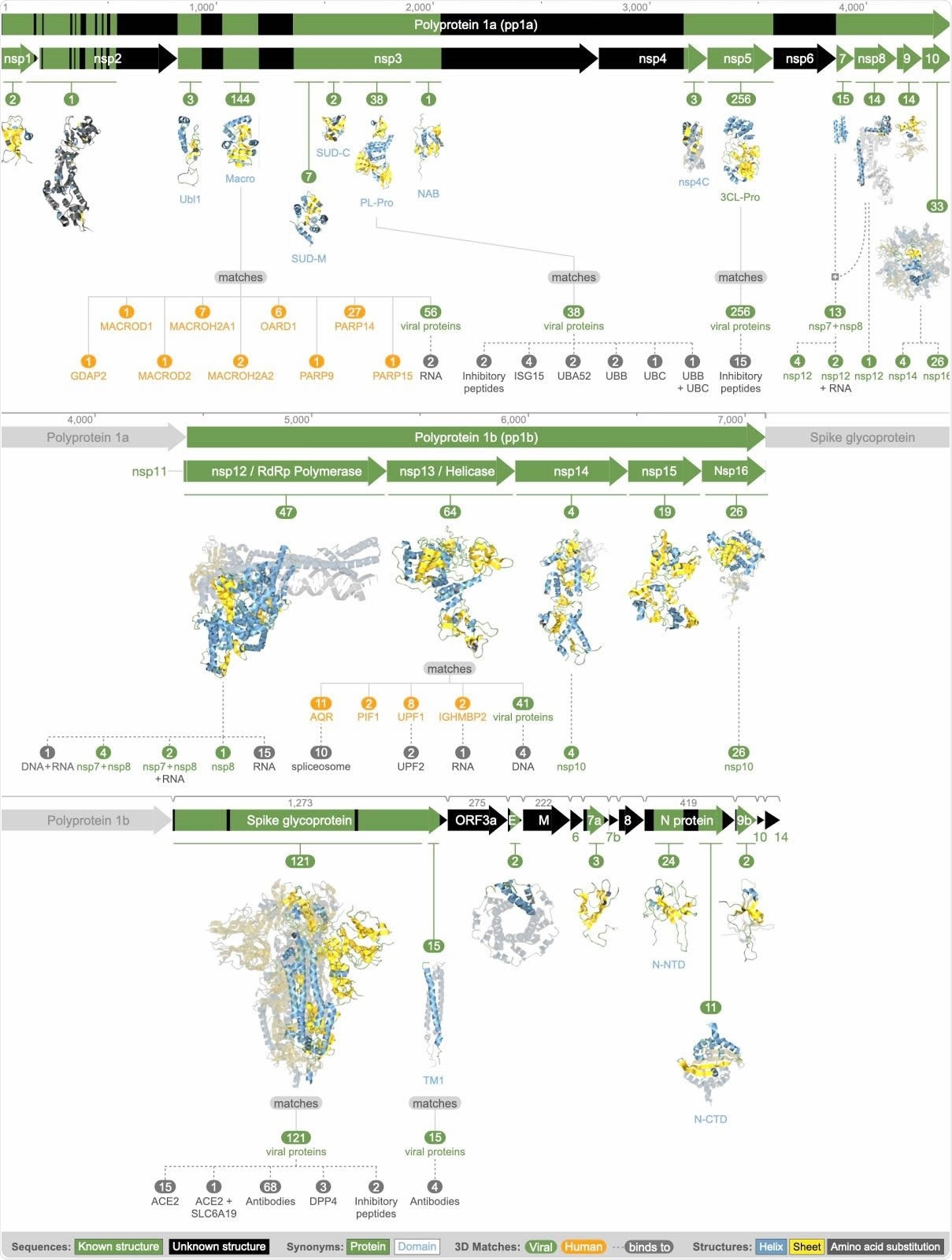
Summary of all available 3D molecular structural knowledge for the viral proteome, as well as derived mimicry, hijacking, and protein interactions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Polyprotein 1a
This comprises non-structural protein (nsps) 1 to 10, with varying degrees of identity to the identified structures, from none, like nsp6, to nsp5, or 3CL-Pro, which was highly conserved and matched 2 CATH families, with 256 matching structures. Another region with multiple matches was the macrodomain region next to nsp2, and part of nsp3, with 144 matches.
Some were highly conserved, but others showed poor conservation.
Polyprotein 1b
The five proteins from this region were all predicted to be highly ordered and to lack transmembrane helices. They include nsp 12 to nsp 16, with as few as four matches for nsp 14 and as many as 64 for nsp 13.
Accessory Proteins and Capsid
The rest of the genome towards the 3’ end, encodes 12 proteins, which are assembled within the cell to form the nucleocapsid. None of these was found to bind to any related 3D structure. The structures with few matches include the ORF3a protein, the envelope protein, the ORF6 to ORF10, and ORF14.
The Envelope Protein
This protein matched two structures from SARS-CoV, one a monomer and one an assembled pentameric protein of five identical units forming a transmembrane ion channel.
Spike Protein
The researchers found 136 matches for this protein in 2 regions, one with 15 structures and matched to the C-terminal transmembrane helix, with four of the matches being with antibody complexes formed against MERS-CoV. The other had 121 matches, 34 almost full-length. These 34 formed a homodimer.
Of the 121, 68 were matched to antibody complexes and another two to complexes with inhibitory peptides. Others were complexed with human proteins, including the ACE2 receptor, with both ACE2 and other human proteins, and to other coronavirus antibodies.
Among all the capsid proteins, this was the only one that had matching structures but was bound to human proteins. The significance of this is that the capsid is mostly assembled within compartments inside the cell, and does not come into contact with host proteins or nucleic acid. There were still four unmatched regions in the region of the spike protein.
Systematic modeling of SARS-CoV-2 protein structures
The Nucleocapsid Protein
This N protein had 35 matches with known 3D structures, in two regions. One region at the N-terminal end contained structures mostly consisting of a single N protein monomer, but with some making up a dimer and one a tetramer. The other, near the C-terminus, had 13 matches, all dimeric structures.
The Value of the Study
The study shows that very few instances of molecular mimicry by the virus of the host proteins were found. Hijacking or self-assembly was also rarely found. In fact, one graph was adequate to represent all such cases, indicating the lack of understanding of the virus’s structural proteins.
Viral Proteins Team Up or Compete
The researchers suggest some interactions between the three proteins in each team.
Team 1 consists of nsp7, nsp8, and nsp12, all of which assemble to produce the viral protein complex in charge of RNA synthesis. Of these, nsp7 was found as a monomer in 2/15 matching structures available at present. Nsp8 was always complexed with nsp12 alone, nsp7 alone, or with both. However, nsp12 was found alone in 38 matching structures.
This finding agrees with older studies on SARS viruses, showing that nsp12 by itself acts as an RNA-dependent RNA polymerase, but shows immense enhancement when it interacts with nsp7 and nsp8. In short, team 1 is characterized by cooperative interactions.
Team 2 comprises nsp10, nsp14, and nsp16. Among the 30 structures that matched either of the latter two, they all had a dimeric structure with one other viral protein, each matching nsp10. Earlier research shows that nsp10 is necessary for the RNA-cap (nucleoside-2′-O-)-methyltransferase activity of nsp14, and for increased activity of 2′-O–-methyltransferase and N-terminal exoribonuclease of nsp16. They conclude that the common region of nsp 10 binds to itself, to form a homo-oligomer, or to nsp14 or to nsp16, in competitive interactions. This could indicate that the amount of nsp10 determines the severity of the infection.
Hijacking Host Cell Functions
Only two viral proteins were found to offer evidence of hijacking of human proteins. The nsp3 PL-Pro may take over the ubiquitin precursors as well as the ubiquitin-like ISG15. This evidence is based on the experimental structures for nsp3 of SARS-CoV, which is known to remove ubiquitin and thus suppress innate immunity.
Another more subtle finding is a much less close match to MERS-CoV, which raises the possibility that this domain may also take over the function of the ubiquitin-60S ribosomal subunit L40. In this way, host ribosomal functions may be hijacked by the SARS-CoV-2 – a hitherto unknown mechanism.
The spike protein hijacks ACE2 as is already established beyond doubt, and this was reflected by 16 structural states showing this complex. Again, a more hidden finding was that it might also take over the cell surface glycoprotein receptor DPP4, promoting T cell activation. This could be one way in which the virus escapes host immunity. The study also revealed 68 matches showing the spike-antibody complex, which could be useful in developing drugs or vaccines that interact with this domain.
Mimicking Host Cell Proteins
The researchers found two human proteins being mimicked by the viral proteins, namely, nsp3 and nsp13. The first is very similar to the macrodomains of nine human proteins, each of which plays one role in the ADP-ribosylation of proteins as a means of a post-translational modification. If so, it can cause epigenomic modification, which could explain why patients show such different responses to the infection. Some of these proteins, like PARP9 or PARP14, modulate macrophage activation as well, which is a known factor in the etiology of vascular disease. Such interactions could explain how a pneumonic illness like COVID-19 progresses to deadly blood vessel damage in many cases.
As for nsp13, a viral helicase, it may mimic any of four human proteins, found to have exon ligation activity, perhaps to create chimeric proteins. By binding to specific other proteins, it could also incite evasion of the cellular immune response.
Suspects
The final group of 17 proteins may be important in viral infection, but structural evidence is lacking. Some had at least one match in known structures, and a few have very well-known roles such as the nsp5, which is the protease cleaving the viral polyproteins. The huge issue is the utter lack of any observations in which the nsp5 or any similar protein interacted with any other host or viral proteins.
The other group had no matching structures at all, and are called structurally dark proteins.
Implications and Conclusion
The researchers comment: “From our analysis, we can conclude that the sequences of these proteins are not detectably similar to any protein that have been observed to date by experimental structure determination methods - at least, based on the sequence-based homology modeling methods used in this analysis.”
They suggest the use of more advanced structural modeling, such as those which use residue-residue contacts along with deep machine learning, to understand the interactions of these proteins in relation to their function in infection.
The study concludes: “Our resource provides researchers with a wealth of information on the molecular mechanisms of COVID-19; the information can easily be accessed, and, to the best of our knowledge, is currently not available at other resources. The resource provides an immediate visual overview of what is known - and not known - about the 3D structure of the viral proteome, thereby helping direct future research.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
O’Donoghue, S. et al. (2020). Systematic Modeling of SARS-Cov-2 Protein Structures. bioRxiv preprint. doi: https://doi.org/10.1101/2020.07.16.207308. https://www.biorxiv.org/content/10.1101/2020.07.16.207308v1
- Peer reviewed and published scientific report.
O’Donoghue, Seán I, Andrea Schafferhans, Neblina Sikta, Christian Stolte, Sandeep Kaur, Bosco K Ho, Stuart Anderson, et al. 2021. “SARS‐CoV‐2 Structural Coverage Map Reveals Viral Protein Assembly, Mimicry, and Hijacking Mechanisms.” Molecular Systems Biology 17 (9). https://doi.org/10.15252/msb.202010079. https://www.embopress.org/doi/full/10.15252/msb.202010079.