A research group headed by Stanford University scientists has developed a scalable, high throughput method to generate high fidelity whole genome and HLA sequencing, viral genomes, and representation of human transcriptome from single nasopharyngeal swabs of coronavirus disease (COVID19) patients. Their results are published on the medRxiv* preprint server.
The ongoing and highly disruptive COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has crippled health care systems around the world and resulted in tremendous morbidity and mortality.
As with other virus outbreaks in the past, viral sequencing has been crucial, although limited by high costs and low throughput. Moreover, the collection of associated host genomic data (which can aid in familial relationship tracking and genetic risk appraisal) has been hampered by the requirement for multiple sampling.
Therefore, there is a pressing need for protocols that can open the door for producing these data in real-time and at scale. This will not only significantly contribute to infection tracking, but also inform further development of therapeutics.
In this groundbreaking study, researchers from Stanford University, Chan Zuckerburg Biohub, University of Lausanne, and Illumina Inc. described a method for achieving simultaneous viral and host sequencing from single SARS-CoV-2 diagnostic nasopharyngeal swab residuals.
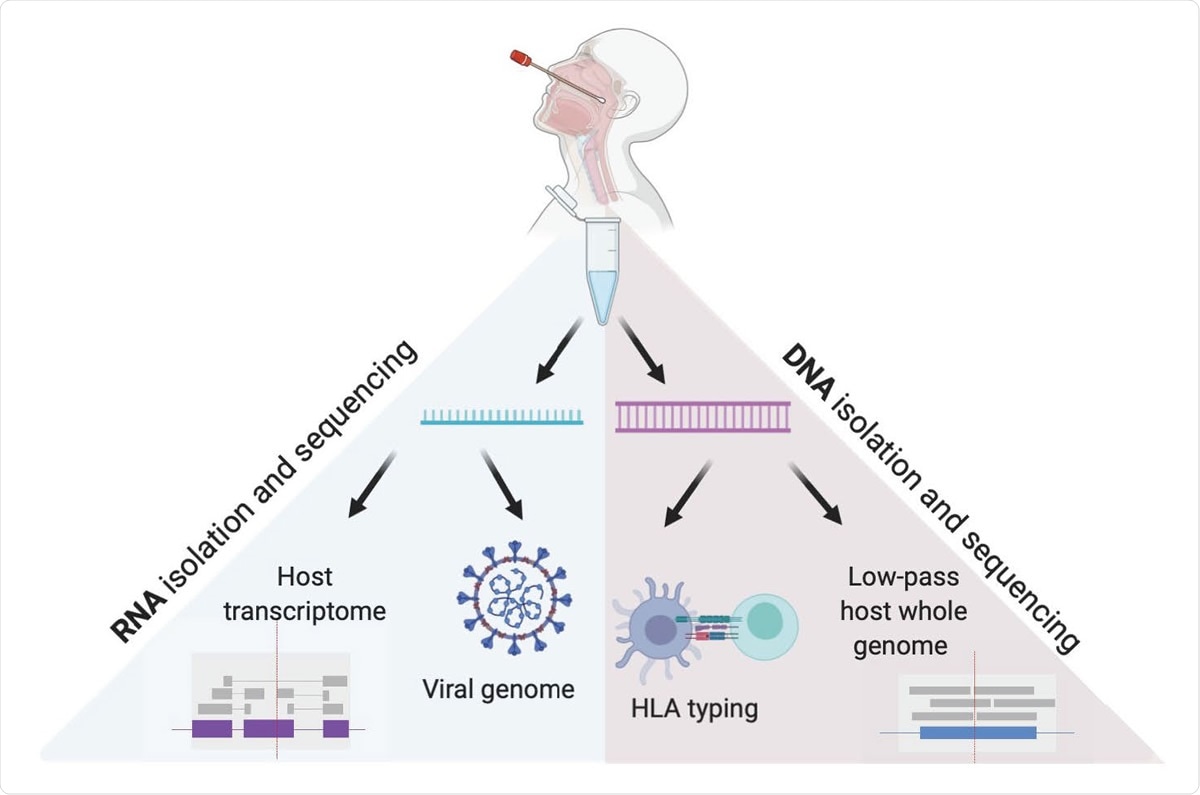
Viral and Host genomes and transcriptomes from a single nasopharyngeal swab. This method allows for independent RNA and DNA isolation from nasopharyngeal swab VTM, enabling viral genome sequencing, detection of host transcriptome, low pass host genome sequencing and HLA sequencing in high throughput.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Analyzing hundreds of samples simultaneously
The researchers used low-pass whole host genome sequencing as an alternative to array-based genotyping in order to provide rich information for trait mapping at scale, which can regularly yield DNA of adequate quality for host genome and HLA sequencing.
Furthermore, they have presented a high-throughput RNA sequencing workflow for sequencing full viral genomes, and human transcriptome reads from hundreds of samples at the same time.
Finally, the researchers described how exactly this method could be used to create a robust multi-omic foundation for data integration and sharing across global institutions – especially since global data repositories have been pivotal for advancing research before and during the current pandemic.
Copious data from a single nasopharyngeal swab
"Here we demonstrate that a single nasopharyngeal swab can reveal substantial host and viral genomic information in a high-throughput manner that will facilitate public health pandemic tracking and research into the mechanisms underlying virus-host interactions", study authors summarize their main findings.
Albeit nasopharyngeal swabs have been used in the past to perform whole-genome sequencing of respiratory viruses in low throughput, this method significantly accelerates the process both in terms of time and number of subjects sequenced.
More specifically, a comparable rate of viral genomic coverage was described, with the capability of studying at least ten times the number of samples in a single sequencing run.
"Using the consensus sequences derived from the initial cohort reported here, as well as samples collected later in March 2020, we created a phylogenetic tree, which allows critical public health phylodynamic tracking", researchers add.
What is also fascinating is that the same nasopharyngeal swab can be used to gather an abundance of human genomics data, and it often yields sufficient DNA to pursue deep sequencing of HLA type, which is a crucial component of the host immune response.
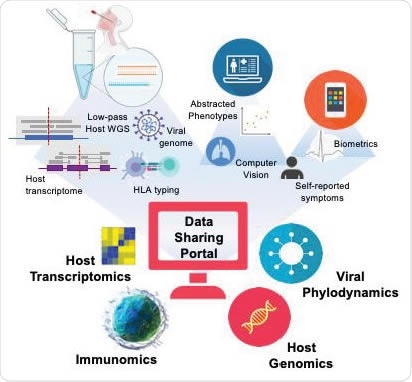
A strong multi-omic foundation for data integration and sharing across global institutions. Using these methods in combination with electronic health record abstraction, and digital medicine, the methods described here builds the foundation for a data repository allowing rapid access to critical data on CoVID19 or any other pandemic via open-source sharing.
The rise of multi-omic data repositories
"Although our initial swab collection did not reveal any viral co-infections, especially as the current pandemic enters the regular flu and cold season, our method allows for acceleration of metagenomics analysis," study authors emphasize the importance of their findings.
Arguably the most significant application of the proposed workflow is that it allows rapid development of large scale, multicentric, and even global host and viral multi-omic data repositories.
With this method, the number of viral genomes comparable to the submissions of SARS-CoV-2 data since the start of the pandemic could be produced by less than a hundred sequencing centers within weeks, along with matched host genome, transcriptome and HLA typing.
And this is basically an indispensable scaffold for integrating such complex inputs to centralized data repositories, enabling, in turn, unparalleled rapidity of the discovery and implementation needed to overcome a devastating COVID-19 pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources