The current COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is known to target primarily the distal lung, including the terminal bronchioles and alveoli, which are the sites of essential gas exchange in the human body.
In a significant minority of patients, this results in critical pneumonia and acute respiratory distress syndrome (ARDS). However, the mechanism by which this occurs is far from clear, and one major contributing factor to this knowledge gap is the absence of a reliable and robust human lung cell culture system that will serve as a substrate for disease of the terminal lungs.
Developing a Robust Long-Term Distal Lung Culture
Now, a new study published on the preprint server bioRxiv* in July 2020 reports the development of a human distal lung culture system that can be functionally tested. This will help not only to understand how this infection produces disease but also to test the proliferative capacity of the stem cells in this part of the body.
As of now, mouse studies provide most of our knowledge about these stem cells, which are functionally part of the lung as well as providing a source of new cells during healing of the lung. These studies have shown that these bifunctional stem cells of the distal lung comprise the secretory club cells found in the distal bronchioles and the type 2 pneumonocytes or alveolar cells (AT2) that produce surfactant in the lung alveoli.
Such studies have also shown that there is both an alveolar progenitor population, which gives rise to both AT1 and AT2 after lung injury, and a basal cell-like or bronchioalveolar progenitor population in the distal airway that give rise to cells in the airway and alveoli. However, are these identical to human lung stem cells?
There are significant differences in the composition of cells within the human lungs relative to the mouse. For one thing, basal cells occur throughout the airway in humans but are not seen in the terminal bronchioles in mice. Instead, club cells carry out renewal and repair processes here in the epithelium. Secondly, the capacity of human AT2 cells to renew themselves long term is unknown as of now.
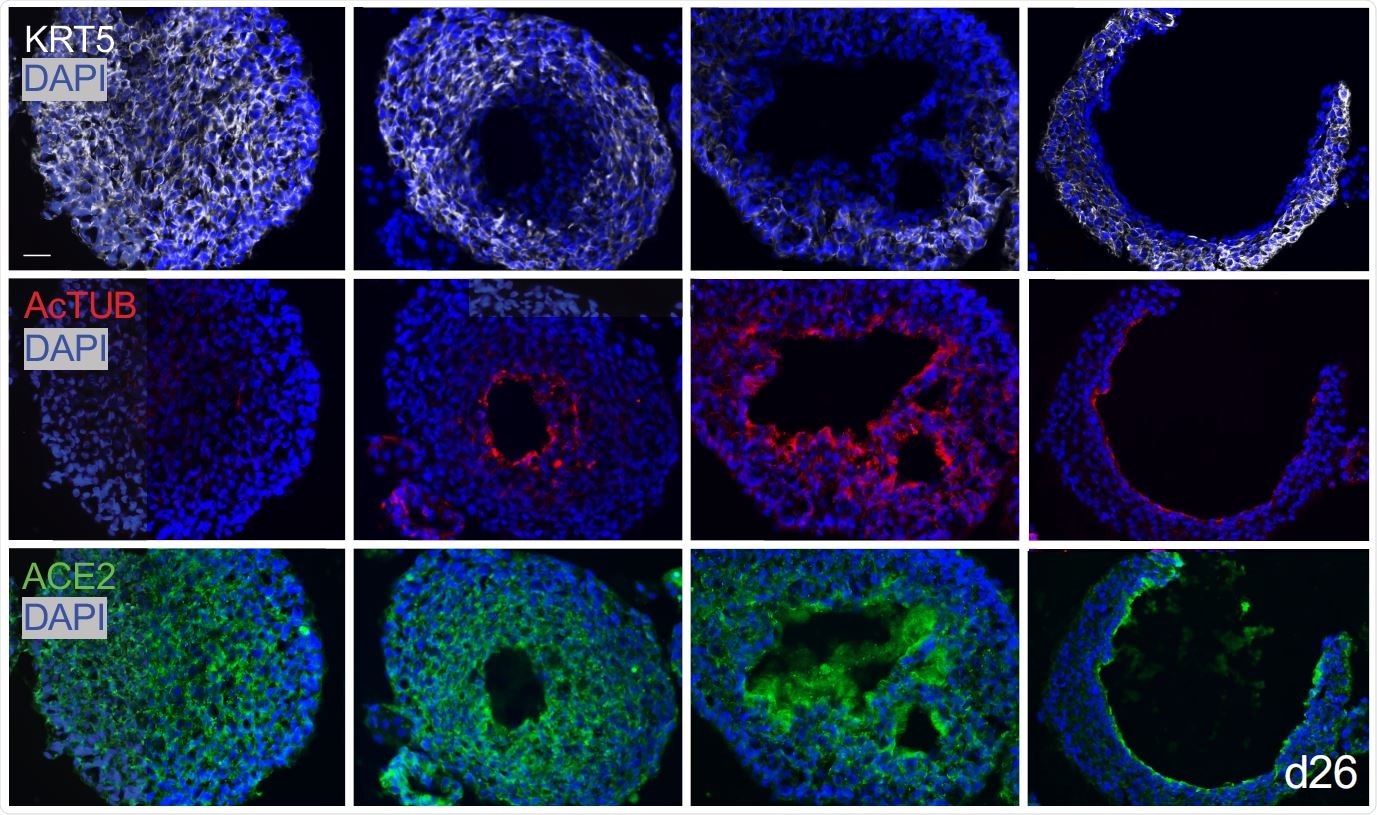
Basal organoids from mixed distal lung culture

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Limitations of Current AT Cell Cultures
While there are several reports of AT cell cultures, these show little expansion due to slow cell turnover. Also, they require feeder cells for their maintenance. This leads to the presence of unknown factors, which could potentially introduce biological variations and makes them less useful for screening purposes. The use of induced pluripotent stem cells (iPSCs) to produce AT2 cells by directed differentiation may also be less useful than expected for the same reasons, and in addition, is inefficient and limited by persistent expression of immature or fetal genes.
Formation of 3D Lung Alveolar and Basal Cell Organoids
The researchers found that in their 3D lung organoids, human AT2 cells renewed themselves extensively, and also were capable of transdifferentiation to AT1 cells. Secondly, they found that basal cell organoids in a mixed culture of distal lung cells first formed solid masses with the KRT5 marker, but after about one month, developed single or sometimes multiple lumens in about half the masses. The luminal cells lost this marker, but other cells, namely, ciliated cells expressing acetylated tubulin and SCGB1A1 markers, merged at the lumen.
Basal 1 Cells Give Rise to Basal 2 Cells
Thirdly, single-cell RNA sequencing of the organoid basal cells with the KRT5 marker from multiple sources showed two subsets of cells, basal 1 and basal 2. The first showed the ability to differentiate as well as markers of cell fate determination, with a subpopulation that showed markers of proliferation. It also had classical lung basal cell mRNA markers like integrin α6, as well as integrin β4. Basal 2 had markers concerned with vesicular and endoplasmic reticulum metabolism as well as squamous cell expression.
The basal 1 subpopulation expressed the marker TNFRSF12A on the cell membrane, which can be used to differentiate a subset of cells enriched for a proliferative gene module, unlike the other markers. Thus, this marker could help pick out human basal cell progenitors.
When the total distal lung organoids were separated by the use of a monoclonal antibody to this marker, they found that cells with TNFRSF12A slowly reduced in number over time, but had a four- to 12-fold higher capacity for clonogenic organoid formation.
Further, FACS analysis showed that basal 2 cells probably originated from basal 1 cells. The basal 1 cells are found to cluster within the distal lung in vivo and have a higher clonogenic potential. In other words, they act like stem cells within the lungs.
Infecting Organoids with SARS-CoV-2 and Influenza Viruses
The researchers were able to establish infection with these viruses in the distal lung organoids, providing proof that these are suited for modeling human infectious disease. Since H1N1 injures both airway and alveolar epithelium, both basal and AT2 organoids were infected by a recombinant H1N1 virus strain, which expressed the fluorescent protein GFP as it replicates. They found that viral genomic RNA built up in the supernatant over 96 hours. These organoids also appeared to possess functional receptors for the influenza virus, similar to those in the intact lung.
When the drug zanamivir was added before the virus, there was no impact on viral replication or infection, because this is a selective inhibitor of viral release from infected cells. However, the nucleoside analog drug Fdc that has been shown to reduce the replication of many viral families was effective. The GFP-expressing virus allowed many antiviral classes of the compound to be screened for differential effectiveness, which indicates this model is useful in the discovery of scalable therapeutics.
Adapting Organoids for Infection by SARS-CoV-2
The current long-term 3D organoids typically have the basal aspect facing the extracellular matrix, or outwards, but this could prevent the infection of the luminal surface, which bears the ACE2 receptors, by the SARS-CoV-2. The researchers, therefore, used an eversion method to generate a robust line of organoids with the apex-out orientation, which increases interactions between the host and the pathogen on the luminal surface.
This resulted in the formation of apex-out spheroids of epithelium where cilia, microvilli, and apical junctions face outwards, accelerating within 5 days to the progressive differentiation of ciliated cells facing outwards over weeks. This is an improvement compared to current protocols of cell culture. These also showed club cells with secretory granules at the outward-facing apex, and the later appearance of AT1 cells in alveolar organoids.
These apex-out mixed organoids were readily susceptible to infection with replication of the SARS-CoV-2 genome at 72 hours from infection being visualized as well as the production of the nucleocapsid protein. This is in contrast to the mechanical shearing required for intestinal organoids to allow apical cell infection with the virus.
About 10% of AT2 cells and basal organoids cells were infected. In the latter subset, the virus was found to infect mostly club cells. This represents a novel target for the virus, which could impair the production of glycosaminoglycans, a protective coating for lung tissue. The loss of this could trigger a vicious cycle of infection with SARS-CoV-2.
Implications and Future Applications
The researchers sum up: “Here, we described a robust, feeder-free, chemically defined method for long-term human distal lung airway and alveolar clonogenic organoid growth, which was applied to progenitor discovery and infectious disease modeling.”
They forecast the application of this method of progenitor culture for all distal lung epithelial progenitors in the adult human to model infectious, interstitial, and neoplastic lung disease, as well as tissue engineering and precision medicine.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources