The spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) depends on its ability to infect new host cells following its replication and release from infected cells. This pivots on its ability to bind to cell surface receptors like the ACE2 enzyme.
Novel Coronavirus SARS-CoV-2 Transmission electron micrograph of SARS-CoV-2 virus particles, isolated from a patient. Image captured and color-enhanced at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Glycosylation of the Spike Protein
The spike protein of the virus is coated with numerous sugar residues, including Asn-linked N-glycans and a few O-glycans linked through Ser or Thr. These glycans allow the protein molecule to offer binding sites for many innate immune cell receptors. These include C-type lectin receptors (CLRs) that bind to specific glycans via the action of calcium.
CLRs are among the abundant immune receptors found on monocytes, dendritic cells, and macrophages, all of which are part of the innate immune system that forms the first line of defense against invading microbes. CLRs are also capable of pattern recognition, especially the one called DC-SIGN, which can marshal host immune responses to target many pathogens via glycan recognition, by their effect on immune system activation induced by Toll-like receptors.
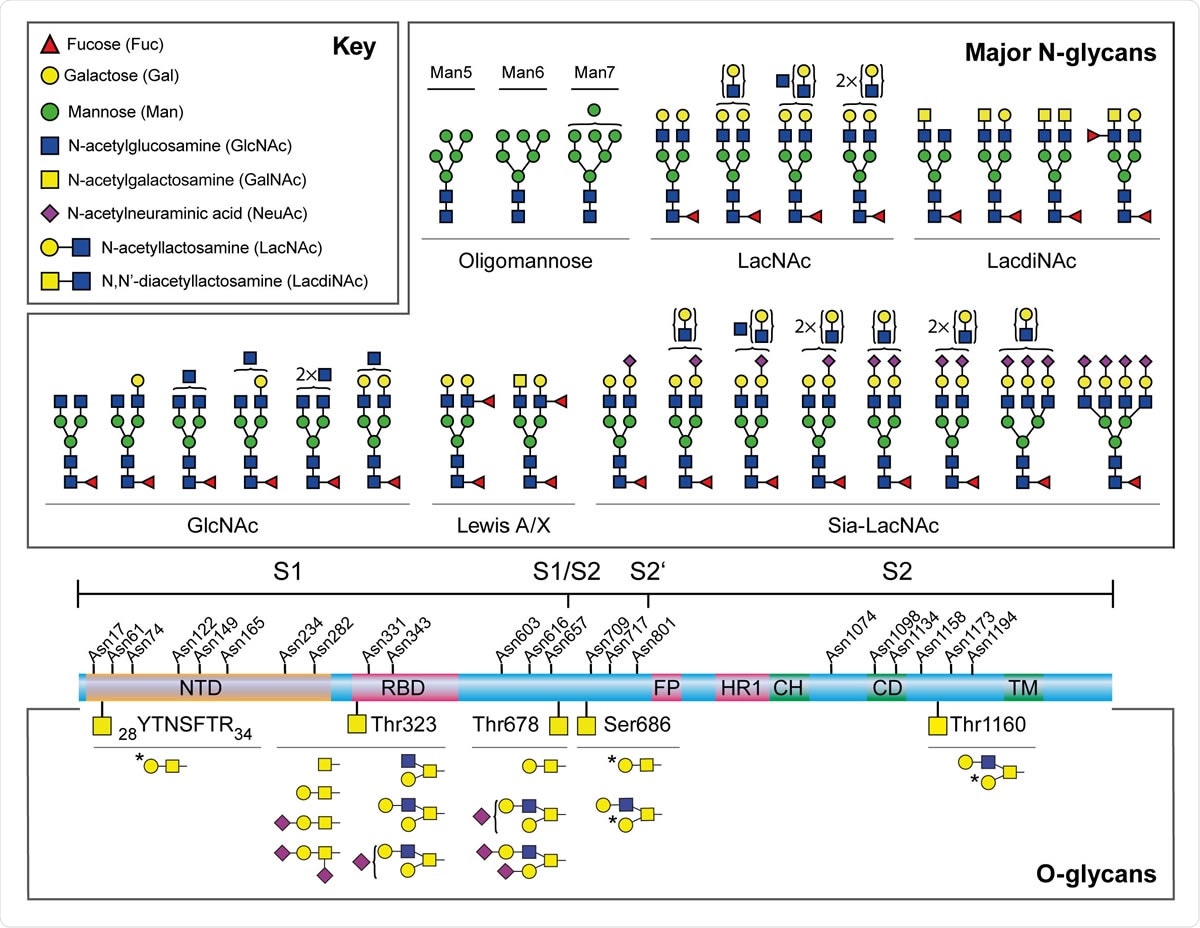
Major N-glycans and O-glycopeptides identified in the recombinant full-length SARS-CoV-2 S. Schematic representation of SARS-CoV-2 S shown in the middle. The positions of N-glycosylation sites are shown on top. Protein domains in the illustration are: N-terminal domain (NTD), receptor-binding domain (RBD), fusion peptide (FP), heptad repeat 1 (HR1), central helix (CH), connector domain (CD), and transmembrane domain (TM). The cleavage sites of S1/S2 and S2’ are labelled. Major N-glycan structures detected by mass spectrometry were categorized by their epitopes on the non-reducing terminal and shown on top. Cartoon symbols above a curly parenthesis indicates sequences corresponding to these compositions cannot be unequivocally defined. The structures presented are only the major glycans on the recombinant full-length S, S1, and S2.
Hyperactive Immune Activity and Glycosylation
Many scientists think it possible that the severe phenotype of COVID-19 is in part due to a hyperactive immune response mediated by innate immune cells. More than 80% of COVID-19 patients show reduced lymphocyte count and a higher proportion of neutrophils to lymphocytes. Their respiratory fluids contain hyperactive macrophages.
Earlier studies show that monocytes and dendritic cells are capable of being infected by SARS-CoV-1, with the DC-SIGN and L-SIGN receptors having the S protein of this virus as their ligand. There were eight glycosylation sites involved in these attachments, and six of these are found in SARS-CoV-2 as well.
The Study: Identifying Virus-Binding CLRs on Innate Immune Cells
The current study was aimed at finding the CLRs that bind via glycan recognition to the S protein of SARS-CoV-2. Some of these receptors bind to oligomannose or to N-glycans on the S protein, as seen by the reduced binding in the presence of other proteins that show the same sugar residues. Others bind to only one type of sugar.
This binding to the viral receptor-binding domain (RBD) is at high affinities measured in picomoles, and in a dose-dependent fashion, though at a lower affinity than the ACE2 receptor. Such binding can, for instance, lead to the recognition of the virus and its capture by the innate immune cells, leading to its internalization and possibly the entry of the S protein into these cells.
The researchers also looked at the N- and O-glycan sequences using recombinant virus particles to pick out those features which determine the interaction of the virus with the CLRs. This analysis shows that while ACE2 receptors are often absent from innate immune cells, CLRs are found in both human tissues and the immune cells found in them. Thus, these could well present new alternative routes for infection.
These receptors could be responsible for the enhanced inflammation and high cytokine levels found in some COVID-19 cases, associated with the presence of monocytes, macrophages, and dendritic cells. Such cells express CLRs abundantly, with almost all macrophages and dendritic cells showing a high level of expression of these receptors.
CLRs like DC-SIGN and MGL were highly expressed in COVID-19 patients with severe disease, who also showed high levels of pro-inflammatory cytokines and chemokines, derived from activated macrophages. These cytokines include IL-6, TNF, CXCL10, and CCL2, and high systemic cytokine levels may be part of the reason for severe COVID-19 infection.
Implications and Future Directions
The researchers say, “Our results offer a possible explanation to how SARS-CoV-2 spreads to extrapulmonary tissues within the host, as the innate immune cells lacking ACE2 expression can still internalize the virus via those CLRs.” The glycosylation of the spike protein is a complex process and depends on both the native protein as well as the infected host cell.
The type and extent of glycosylation will, therefore, determine the receptor-virus interaction in innate immune cells, causing either viral clearance or viral spread to other organs and other hosts. The virus internalization observed following CLR binding could lead to a slower onset of symptoms by reducing the titer of free virus, and this may be one reason for the asymptomatic or presymptomatic infection.
However, HIV studies show that such binding can also enhance trans viral transmission, especially with cells that express DC-SIGN and L-SIGN, since these cells have been shown to transfer SARS-CoV to vulnerable target cells. Again, DC-SIGN expression is strongly linked to the genes that determine the blood group, which is a genetic factor that affects the risk of respiratory failure in COVID-19.
The researchers speculate, “Subsequent to CLR-dependent internalization, SARS-CoV-2 could be conveyed by CLR-expressing innate immune cells, and redistributed to permissive tissues where more damage could occur.” In fact, a recent study showed the presence of viral nucleocapsid protein in the spleen and lymph nodes.
The study shows that small molecules that bind the virus, or which prevent the spike glycans from interacting with the CLRs, could reduce the extent of virus distribution in these patients as well as restricting hyperactive immune responses. The genetic basis for CLR variation could show links between CLRs and COVID-19 severity. The role played by infected immune cells in distributing the virus is also to be studied. At the same time, the variation in S glycosylation could also help explain differences in disease phenotype and virus spread.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.